8.7 Molecular similarity
In order to analyze the similarity between
molecules, VEGA
ZZ can superimpose them and calculate the RMS (Root Mean Square) value. This function is available choosing
Calculate
![]() Similarity in the main menu.
Similarity in the main menu.
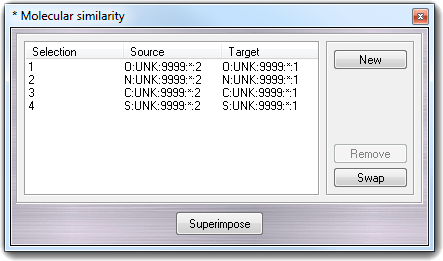
To superimpose two molecules, you must
click at least three atom pairs.
Remember that the source molecule is moved on the target molecule and the source
is defined at first atom click. If you want swap the source with the target, you
must click Swap button. To remove a single pair, you must
select it in the list and than click the Remove button. To restart the
selection of the atom pairs, you must click the New button. To perform
the superimposition of the two molecules, you must click the Superimpose
button and to revert the source molecule to the starting position after the
superimposition, use the undo function.
When you perform the superimposition, the RMS value is reported in the VEGA ZZ
console.
8.7.1 The context menu
Clicking the right mouse button on the selection list, the context menu is shown. New, Remove and Swap have the same functions of the correspondent buttons and Selection sub-menu allows to do automatic selections without to click the atom pairs. Use All pairs to select all possible atom pairs. This feature is useful to compare conformers each other, because they are the same molecule. Similar pairs allows to select atom pairs that have atom name similar to highlighted pair. This is useful to select the backbone of a protein.
Checking Active only, you can automatically select the atom pairs of active/visible atoms only and you can also define the search direction along the atom list by checking Ascending, Descending and Both in Direction sub-menu.