12.14 PropKa - Prediction of protein pKa values
12.14.1 Introduction
Hui Li, Andrew D. Robertson and Jan H.
Jensen developed a very fast empirical method for protein pKa
prediction and rationalization (PROTEINS: Structure, Function and Bioinformatics
61:704-21, 2005). The desolvation effects and intra-protein interactions, which
cause variations in pKa values of protein ionizable groups, are
empirically related to the positions and chemical nature of the groups proximate
to the pKa sites. The Authors developed a program (PROPKA) to
automatically predict pKa values based on these empirical
relationships within a couple of seconds. Unusual pKa values at
buried active sites, which are among the most
interesting protein pKa values, are predicted very well with the
empirical method.
12.14.2 Usage
PropKa plug-in requires a
pre-loaded molecule in VEGA ZZ workspace and it works with proteins only. To
start the prediction, you must select Calculate
![]() Protein pKa
in main menu. The text output generated by PROPKA is shown in VEGA ZZ
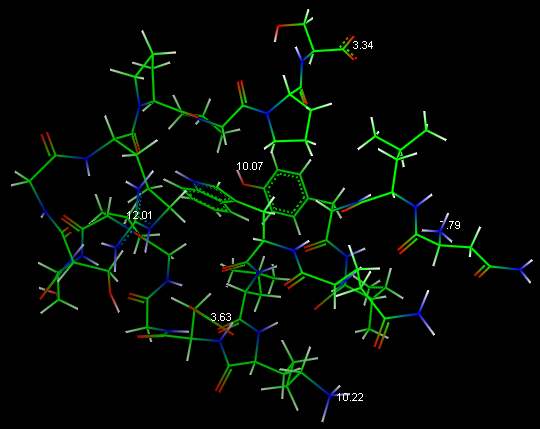
console and the pKa values in the 3D graphic environment:
Protein pKa
in main menu. The text output generated by PROPKA is shown in VEGA ZZ
console and the pKa values in the 3D graphic environment:
Example of console output:
-----------------------------------------------------------------------------------------------
-- PROPKA: A PROTEIN PKA PREDICTOR --
-- VERSION 1.0, 04/25/2004, IOWA CITY --
-- BY HUI LI --
-- --
-- PROPKA PREDICTS PROTEIN PKA VALUES ACCORDING TO --
-- THE EMPIRICAL RULES PROPOSED BY --
-- HUI LI, ANDREW D. ROBERTSON AND JAN H. JENSEN --
-----------------------------------------------------------------------------------------------
------- ----- ------ -------------------- ------------- ------------- -------------
DESOLVATION EFFECTS SIDECHAIN BACKBONE COULOMBIC
RESIDUE pKa LOCATE MASSIVE LOCAL HYDROGEN BOND HYDROGEN BOND INTERACTION
------- ----- ------ --------- -------- ------------- ------------- -------------
GLU 1 4.50 SUFACE 0.00 82 0.00 0 0.00 000 0 0.00 000 0 0.00 000 0
ASP 9 2.76 SUFACE 0.00 147 0.56 8 -0.80 LYS 8 0.00 000 0 0.00 000 0
ASP 9 -0.80 ARG 16 0.00 000 0 0.00 000 0
TYR 3 10.00 SUFACE 0.00 142 0.00 0 0.00 000 0 0.00 000 0 0.00 000 0
N+ 1 7.51 SUFACE 0.00 117 -0.49 7 0.00 000 0 0.00 000 0 0.00 000 0
LYS 8 9.94 SUFACE 0.00 145 -0.56 8 0.00 000 0 0.00 000 0 0.00 000 0
ARG 16 12.08 SUFACE 0.00 143 -0.42 6 0.00 000 0 0.00 000 0 0.00 000 0
-----------------------------------------------------------------------------------------------
SUMMARY OF THIS PREDICTION
ASP 9 2.76
GLU 1 4.50
TYR 3 10.00
N+ 1 7.51
LYS 8 9.94
ARG 16 12.08
-----------------------------------------------------------------------------------------------