| 3.2 Force Field Attribution |
VEGA allows to attribute the force fied potential using a new description language (Atom Type Description Language, ATDL) to recognize the atom types. VEGA can also assign the atomic charges using the
Gasteiger-Marsili method (5) or a fragment database.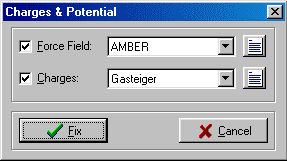
Charges & Potential dialog box
The atom typing is centred to the assumption that atoms in analogous chemical environment will usually have local properties that are transferable between molecules. This concept plays a pivotal role not only in the parameterization of the potential energy functions (force field), but also in the calculation of all molecular properties based on an atomic parametrization (e.g. logP prediction).
For this reason we sought a method able to describe the atom typing of both force fields and atomic parametrizations. We here propose ATDL language that is able to define any atom type and it makes possible to change from one atom type classification to another, referring to the corresponding ATDL definition.
ATDL is based on the molecular connectivity and each atom is defined describing not only the directly bonded atoms, but, using the recursive character of C language, it’s possible to define theoretically infinite sublevels of connected atoms. This characteristic allows a more accurate definition of atom types and reduces the conflicting assignments. The user can expand VEGA capabilities, writing new force field templates with ATDL.
3.2.1 Algorithm Details
The recognition and the assignment of correct atom types using ATDL language is made up of three step. Firstly VEGA determines the atomic connectivity. To make it VEGA doesn’t use existing information (e.g. CONECT records from PDB files), but resolves it, calculating all interatomic distances using a table of Van der Waals radii with a tolerance threshold equal to 20%. From the connectivity VEGA determines also the bond orders and the hybridization types of all atoms. VEGA can handle only atoms with filled valencies (i.e. with all hydrogens) and so VEGA doesn’t consider the valence angle, but only the bond length.
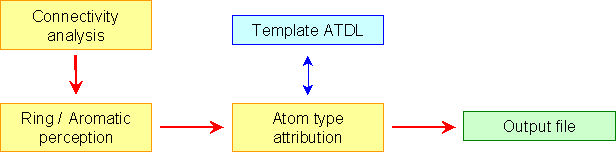
In the second step, VEGA finds the ring systems. VEGA looks for the rings from three to seven member by means of an algorithm that analyzes recursively the connected atoms for all atoms and determines the closed connections. The perception of aromatic rings is based on Hückel’s rule. A ring is classified as aromatic if it has 4n + 2 ¶-electrons. All bonds within the ring are typed as aromatic and all hybridization states are sp2. VEGA recognizes both aromatic and heteroaromatic rings with five or six atoms.
Finally VEGA checks the correct syntax of the chosen template file with a description parser and matches the examined atom with the ATDL descriptions to find the corresponding atom type.
3.2.2 ATDL Specifications
Each atom is defined by a five-character string. The first two characters are the element symbol of atom. If the element symbol is one character only, the second character must be a dash (-). For a better description, special elements are included: X for any atom and # for any heavy atom. The third character is the bond order: one can use values from 1 to 6 for real bond order, 0 for non bonded atom and 9 for a bonded atom without specified bond order. The fourth character is the ring indicator: ranging from 3 to 6 if the atom is a 3 to 6-ring member, 0 for a non-ring member atom and 9 for a non-specified ring atom. The fifth character is the aromatic indicator: 0 for non-aromatic atom, 1 for aromatic atom and 9 if the aromaticity is non-specified.
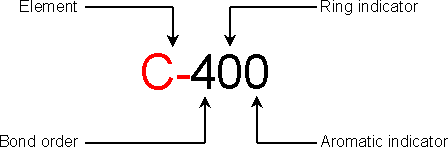
Examples:
C-300 is a sp2 aliphatic carbon, Si900 is a generic silicon with any bonds
order and C-361 is an aromatic carbon included in a six-member ring.
After the atom definition you can write the description of bonded atoms (also in ATDL) limited by parenthesis as the description of connected atoms is crucial to correctly define the atom properties. For example O-200 indicates an oxygen atom not involved in a ring bonded to two other atoms. This specification can be used both for an hydroxyl group and for an ether function. In order to resolve this disagreement you can point out the description of connected atom in ATDL: O-200 (C-400 H-100) for hydroxyl group and O-200 (C-400 C-400) for ether.
More than one level of parenthesis can be used for complex description of atom types and each group of atoms, limited by parenthesis, contains the descriptions of all atoms bonded to the previous atom.
For instance the description of the
a carbon atom of a glycine in zwitterionic form is:C-400 (N-400 (H-100 H-100 H-100) C-300 (O-100 O-100) H-100 H-100)
In this way you can indicate a sp3 carbon bonded to two hydrogen atoms (H-100), a nitrogen atom positively charged (N-400) with three hydrogen atoms and a sp2 carbon bonded to two oxygen atoms non substituted.
With ATDL specifications it’s virtually possible define any atom type describing all bonded atoms until giving a representation of whole molecule.
For each atom typing scheme (force field or atomic parametrization) VEGA uses a template file, in which the descriptions of atom types are stored along with a symbolic code (for force fields) or a local value (for atomic contributions). Each atom type definition is stored in a line with three columns. The first column is the atom type name (or value), the second column is the atom description in ATDL and the third column contains the descriptions of bonded atoms (also in ATDL).
Actually VEGA supports several force fields, e.g. CVFF (6), AMBER (7), CHARMM (8), MMFF (9) and TRIPOS (10), but everybody can expand VEGA with new atom type descriptions or new template files.
| 3.3
Molecule Analysis |