12.10 Pockets - Protein cavity detection and mapping
12.10.1 Introduction
Pockets is the graphic interface for fpocket, the well known software developed by Vincent Le Guilloux and Peter Schmidtke to detect protein cavities. This program uses an extremely optimized algorithm based on Voronoi tessellation whose performances allow to analyze large molecules in few seconds.
The Pockets plug-in was developed with the intent to integrate fpocket into VEGA ZZ, making easier the use of this command-line oriented software. The possibility to identify protein cavities is very useful to perform blind docking studies when the binding site is unknown. Fpocket with Pockets plug-in helps you to scan the target protein to find the best pockets able to accommodate ligands, whose binding mode is not yet identified. Moreover, the last version of the plug-in can map the cavities detected by fpocket performing a docking calculation of one or more probe molecules with AutoDock Vina or PLANTS docking programs. For a better ranking of the cavities with both fpocket and docking scores, Pockets can calculate also a consensus score according to a set of parameters selected by the user.
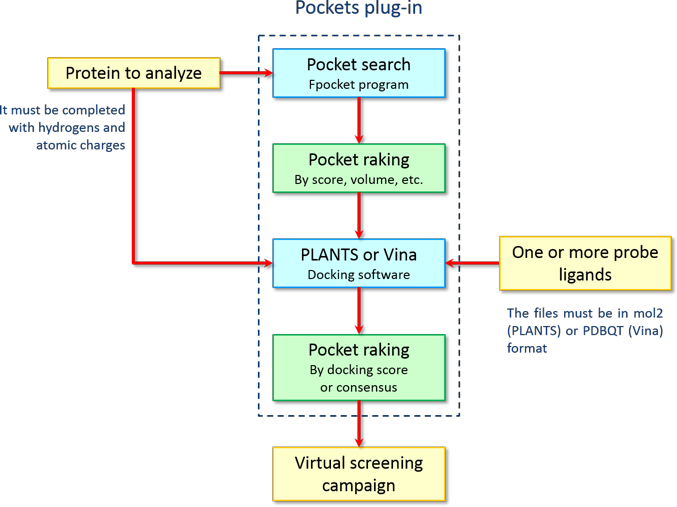
Therefore. the workflow for the identification of the potential binding site of a protein is summarized in the following chart:

By this protocol, it is possible to identify the most probable binding sites according on both physicochemical properties and the capability to bind ligands of the pockets without the need to perform a full blind docking, which is affected by intrinsic inefficiency of docking algorithms when have to explore a large area of the target protein. In this way, it is possible to improve the probability of success in finding the right binding site, reducing dramatically the computational time. Thank to the parallel execution of the docking calculations, it is possible to complete a full analysis of the pockets (pocket search, docking calculation and ranking) in few minutes.
12.10.2 Pocket search
Before to start the pocket analysis, you must open the protein to study,
which must be completed with hydrogens (see
Edit
![]() Add
Add
![]() Hydrogens) and the atomic charges must be correctly assigned if you want to
perform the pocket mapping with docking calculations (see
Calculate
Hydrogens) and the atomic charges must be correctly assigned if you want to
perform the pocket mapping with docking calculations (see
Calculate
![]() Charge & Pot.). After these operations, the Pockets main window can be
shown bt selecting Calculate
Charge & Pot.). After these operations, the Pockets main window can be
shown bt selecting Calculate
![]() Pockets in
the VEGA ZZ main menu:
Pockets in
the VEGA ZZ main menu:
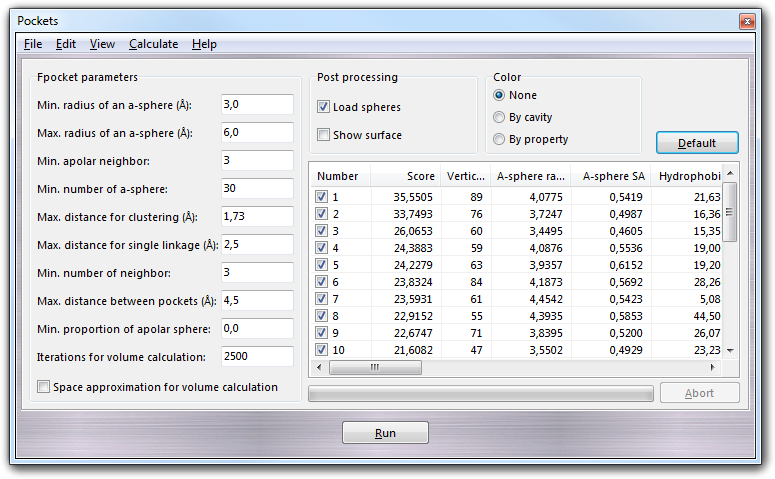
In Fpocket parameters box, you can set all parameters to control the cavity search and, for an exhaustive explanation, you can read the fpocket manual. In Post processing box, it's possible to define the actions when the fpocket calculation is finished: checking Load spheres, the spheres, calculated by fpocket to fill the cavities, are automatically loaded in the 3D graphic window of VEGA ZZ and checking Show surface, the cavity surfaces are calculated and shown. In Color box, you can define the coloring methods of cavities: None (default color only), By cavity (each cavity has a different color) and By property (the surface is colored by polar (violet) and apolar (sand) properties).
Clicking Run button, a file requester is shown asking you the PDB file needed as input by fpocket and clicking Save button, the fpocket calculation starts. At the end, the results are automatically loaded in the table as shown above. The cavities are sorted by score and the meaning of each column is explained in the fpocket manual. Additional columns are added by the plug-in, which are very useful to setup a docking calculation, as shown in the following table:
| Column name | Description |
| Center X | The Cartesian coordinates of the centre of the pocket. |
| Center Y | |
| Center Z | |
| Radius | The radius in ┼ of the approximate sphere including the pocket. |
| Size X | The approximate size along X, Y and Z axis of the pocket. |
| Size Y | |
| Size Z |
The cavities can be sorted also in ways other than the predefined one (in ascending or descending order) just clicking the header of the column corresponding to score/property by which you want to sort. To show the spheres filling a specific pocket or their surfaces, you can double click the corresponding row in the result list. Clicking the table by the right mouse button, the context menu is shown and its features are summarized in the following table:
| Menu | Subitem | Description |
| New | - | Clear the table of the results. |
| Open | - | Open the cavities previously calculated. You can select the *_out or the pockets directory. If the current workspace is empty, you can't show the surfaces double clicking on them. |
| Save as ... | - | Save the pocket table in Comma Separated Values (CSV) format or in Data Interchange Format (DIF). |
| Show | Spheres | Show the pocket by sphere or surface. |
| Residues | Show the protein residues facing in the cavity. | |
| Probe pose |
Show the probe pose if you performed a docking calculation. You must click with the right mouse button on the cell of the Probe column corresponding the ligand that you want to show. That's is needed because you can map the pockets with more than one probe. |
|
| Select | Top | Select (check) the top ranked pockets (1, 3, 5, 10, 20, 50 and 100). |
| All | Select all pockets. | |
| None | Unselect all pockets. | |
| Copy | All | Copy the whole table to the clipboard in CSV format. |
| Value | Copy the value under the mouse pointer to the clipboard. | |
| Checked | Copy only the selected/check pockets/rows to the clipboard. | |
| Row | Copy the row to the clipboard. The values are separated by space characters. | |
| Column | Copy the column to the clipboard. If you show the context menu clicking with the right mouse button on the column header, also the label is copied, otherwise it is missed. The values are separated by line feed characters. | |
| Send to Excel | All | Send the complete table of the results to Microsoft Excel through the DDE connection. |
| Checked | Send only the checked/selected rows to Microsoft Excel. | |
| Remove pockets | - | Remove all pockets from the current VEGA ZZ view. |
| Remove column | - | Remove column and only Probe and Consensus column can be removed. You must pay attention because when you remove the Probe column also the docking data are deleted (moving it to the trashcan if the data are on a local drive). A requester ask you if continue or not when you are removing a Probe column. |
Some of these functions are duplicated in the menu bar, while others are unique as shown in the following table:
| Menu | Submenu 1 | Subitem 2 | Description |
| File | Open | - |
Open the cavities previously calculated. You can select the *_out or the pockets directory. If the current workspace is empty, you can't show the surfaces double clicking on them. |
| Save as ... | - | Save the surface table in Comma Separated Values (CSV) format or in Data Interchange Format (DIF). | |
| Close | - | Close the plug-in window. | |
| Edit | Copy | Checked | Copy only the selected/check pockets/rows to the clipboard. |
| Row | Copy the row to the clipboard. The values are separated by space characters. | ||
| All | Copy the whole table to the clipboard in CSV format. | ||
| Send to Excel | All | Send the complete table of the results to Microsoft Excel through the DDE connection. | |
| Checked | Send only the checked/selected rows to Microsoft Excel. | ||
| View | Select | Top | Select (check) the top ranked pockets (1, 3, 5, 10, 20, 50, 100). |
| All | Select all pockets. | ||
| None | Unselect all pockets. | ||
| Remove pockets |
- |
Remove all pockets from the current VEGA ZZ view. | |
| Calculate | Pockets | - | Run fpocket to calculate the cavities. It's equivalent to press the Run button. |
| Docking | - | Perform the mapping of the pockets by docking (see below). | |
| Consensus | - | Calculate the consensus of a user-defined set of scores (see below). | |
| Help | Manual | - | Show this manual. |
| Fpocket | - | Show fpocket manual. | |
| About | - | Show the copyright message. |
Clicking the Default button, you can revert to default settings.
Notes:
All fpocket executables (dpocket, fpocket and tpocket) are included in VEGA ZZ package and can be used also without the GUI as stand-alone programs based on command-line input.
12.10.3 Docking calculation
Before to to perform the docking calculation, you must:
About the last point, PLANTS doesn't need special precaution in
preparing the probe if not the file format that must be mol2 and atom
charges must correctly assigned. On the contrary, Vina requires several
operations to prepare the ligand to dock and, for this reason, it is strongly
recommended to use the script Docking
![]() Vina
Vina
![]() Ligand.c to do automatically all needed operations (e.g. charge assignment,
atom type assignment, deletion of apolar hydrogens, search for flexible torsions
and save in PDBQT format).
Ligand.c to do automatically all needed operations (e.g. charge assignment,
atom type assignment, deletion of apolar hydrogens, search for flexible torsions
and save in PDBQT format).
When you have completed all these steps, you can select Calculate
![]() Docking in the plug-in menu bar and than the docking dialog window will be
shown:
Docking in the plug-in menu bar and than the docking dialog window will be
shown:
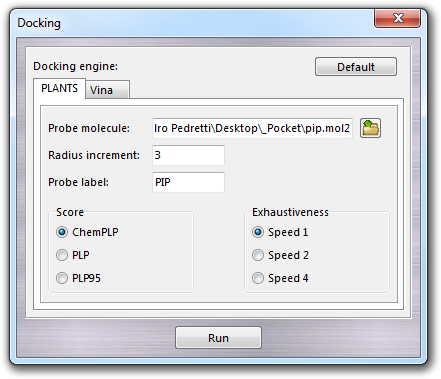
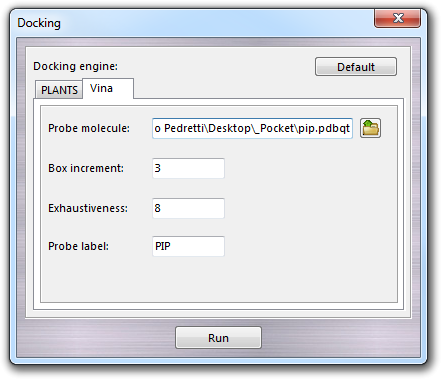
Here you can select the docking engine (PLANTS or Vina) and set the main parameters such as the Probe molecule file previously prepared as described above and the Probe label, which is used as header of table column of the docking scores. The label is automatically proposed by the program, but it can be changed by the user according the following rules: 1) it must written with capitalized characters; 2) only alphanumerical characters are admitted (no colons, commas, dashes, dots, underscores, etc.); 3) the maximum length is 8 characters. Other specific parameters must be set for each docking engine.
12.10.3.1 PLANTS parameters
When you select PLANTS as docking engine, you must set the Radius increment whose value is added to the radius of the sphere including the pocket, the Score (ChemPLP, PLP and PLP95) and the Exhaustiveness level of the calculation. In particular, Speed 1 is the slowest level, but it is best in accuracy. Speed 4, as reported in PLANTS manual, is the fastest but is not recommended and is present only for test purposes.
WARNING:
PLANTS is not included in VEGA ZZ package, but is available for
free for non-profit institution and can be requested to its owners.
12.10.3.2 Vina parameters
AutoDock Vina requires to set different parameters such as the Box increment, which is the value added to the X, Y, Z dimensions of the box including the pocket and the Exhaustiveness of the calculation. Vina program is included as both 32 and 64 bit versions in VEGA ZZ package and doesn't need to be installed.
The Default button reverts the settings to default, while the Run button starts the docking, which can be aborted in any time by clicking the Abort button in the Pockets main window. Multiple instances of the docking programs are started simultaneously according the total number of cores/threads of the CPUs in order to maximize the performances. Only the checked pockets are selected for the docking and in this way you can reduce the calculation time selecting only the best ranked pockets.
At the end of the docking, a column named Probe XXX, where XXX is the label of the probe is added to table of the result.
12.10.4 Consensus calculation
As final step, to rank the pockets according to both pockets
scores/properties and docking scores, you can calculate the consensus score and
order by it. In particular, selecting Calculate
![]() Consensus, the following dialog is shown:
Consensus, the following dialog is shown:
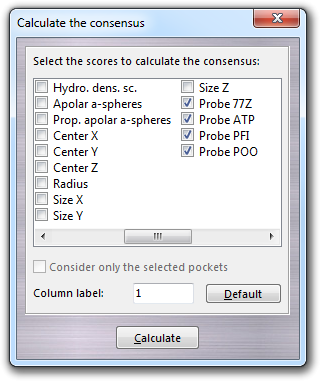
Here it is possible to combine each other docking scores (labelled as Probe XXX) with pocket scores (e.g. the Score column, not shown in the above picture). In this dialog, you can also specify to calculate the consensus only for the checked/selected pockets and the label of the column, which is added to the list of the results. The same rules for the probe label are valid also here. Clicking the Default button, all parameters are reverted to default and in particular Score and Probe XXX columns are automatically selected. Finally you can sort the pocket according to the consensus (lower = better) by clicking with the left mouse button on the header of the column, which is named as Consensus XXX, where XXX is the label indicated in the Column label field. The consensus columns are not automatically saved, but can be exported to Excel with the other data or copied to the clipboard.
12.10.5 Output files
All files are saved in the output directory generated by fpocket, whose name is the same of the PDB file needed as input (PROTEIN) followed by _out suffix. In details, the following scheme shows its structure:
| PROTEIN_out | Root of the output directory |
|
Directory of the pocket files (two files for each pocket enumerated from 0). |
|
Atoms defining the border of the pocket #0 in PDB format. |
|
Spheres filling the pocket #0 in PQR format. |
|
|
|
Directory the best poses for each pocket. If more than one probe was used for the analysis, this multiple copies of this directory could be present with different PROBEID. |
|
Best pose of the probe for the pocket #0 in mol2 format. This kind of file is generated if you selected PLANTS as docking engine. |
|
Best pose of the probe for the pocket #0 in PDBQT format. This kind of file is generated if you selected Vina as docking engine. |
|
Script to show the pockets whit PYMOL. |
|
Script to show the pockets whit VMD. |
|
PDB file including both input protein and all pockets. |
|
Spheres of all pockets in PQR format. |
|
PLANTS output file with all docking scores in CSV format (one for each probe). |
|
Vina output file with all docking scores in CSV format (one for each probe). |
|
Batch file to run PYMOL. |
|
Batch file to run VMD. |
12.10.6 Copyright
fpocket
is a software developed in 2009-2021
by Peter Schmidtke, Vincent Le Guilloux and Pierre Tuffery
All rights reserved.
AutoDock Vina
is a software developed in 2010-2021
by Oleg Trott
All rights reserved.
PLANTS
is a software developed in 2006-2021
by Oliver Korb, Thomas StŘtzle and Thomas Exner
All rights reserved.
Pockets plug-in
for VEGA ZZ
is a software developed in 2010-2021
by Alessandro Pedretti and Giulio Vistoli
All rights reserved.
Alessandro Pedretti
Dipartimento di Scienze Farmaceutiche
FacoltÓ di Scienze del Farmaco
UniversitÓ degli Studi di Milano
Via Luigi Mangiagalli, 25
I-20133 Milano - Italy
E-Mail: info@vegazz.net