To open the main window, you must select Bioinformatics
12.13 Predator - Secondary Structure Prediction
This plug-in interfaces VEGA ZZ to Predator* that allows
to predict the secondary structure of a protein starting from their primary structure. It
takes as input a single protein sequence to be predicted and can
optimally use a set of unaligned sequences as additional information to predict the query
sequence. The mean prediction accuracy of PREDATOR is 68% for a single sequence and 75%
for a set of related sequences.
To open the main window, you must select Bioinformatics
![]() Predator
from the main menu.
Predator
from the main menu.
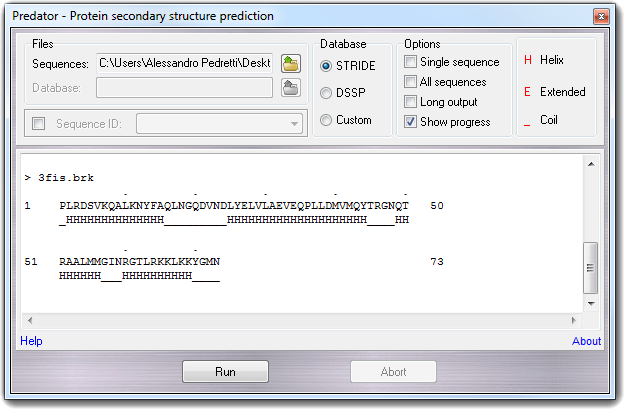
In Files box, you can enter the file containing on or more sequences according to Predator's
supported files (in FASTA, CLUSTAL,
MSF formats) and the file name of the custom database.
Checking Sequence ID box, you can select the sequences that Predator uses as
query. The default sequence is the first in the sequences file. Database box allows to select the database used to predict the secondary
structure. If Custom is selected, you can insert the custom database
file name in Files box. In Options box, you can change some Predator parameters:
Pressing Run button, the calculation starts and can be stopped clicking on Abort button. The output is directly captured in the big edit box and it's shown as below:
Info> 3fis.brk : .................................................!
Info> FIS_HAEIN : .................................................!
Info> NTRC_AZOBR : .................................................!
Info> NTRC_RHIME : .................................................!
Info> NTRC_BRASR : .................................................!
Info> NTRC_RHOCA : .................................................!
Info> ATOC_ECOLI : .................................................!
Info> NTRC_KLEPN : .................................................!
Info> FLBD_CAUCR : .................................................!
Info> NTRC_ECOLI : .................................................!
Info> NTRC_SALTY : .................................................!
Info> NTRC_PROVU : .................................................!
Info> Identical 7-residue fragments found in:
Info> 3fis.brk B
> 3fis.brk
. . . . .
1 PLRDSVKQALKNYFAQLNGQDVNDLYELVLAEVEQPLLDMVMQYTRGNQT 50
_HHHHHHHHHHHHHH_________HHHHHHHHHHHHHHHHHHHH____HH
. .
51 RAALMMGINRGTLRKKLKKYGMN 73
HHHHHH___HHHHHHHHHH____
Clicking Help label, this document is shown, and clicking About
label, the following copyright dialog appears:
For more information about Predator software, see the original documentation:
*Predator is copyrighted by Dmitrij Frishman & Patrick Argos