| Drug Design Laboratory |
|
 |
| Sitemap |
| Print Version |
| Login |
| About CMSimple |
| News > Software projects > Tree2C |
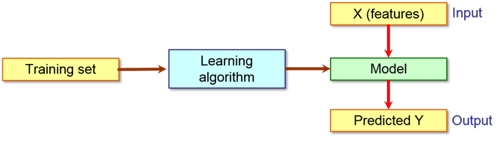
Tree2CClassification tree to code converter
This program converts the machine learning models, in particular the classification trees, generated by Weka program mainly to C source code but it supports also other programming languages (e.g. C++, Fortran 90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL and VBScript). The resulting code requires no or very limited modifications to be used. The program can recognize several molecular attributes/descriptors (especially those that are calculated by VEGA ZZ and MOPAC 2016) and it can add automatically the code to calculate them. Moreover, Tree2C can generate code also for the domain property check, which is a very useful feature to evaluate the confidence of the classification results. In addition, the program can build the code for different targets to be integrated in a pre-existing program:
For more information, you can consult the manual System requrements: Tree2C supports both Linux (x86 or x64) and Windows (2000/XP/Vista/7/8/8.1/10 x86 or x64) operating systems and requires the HyperDrive runtime library, which is the same used for VEGA ZZ software. Since the program is written in standard C code, it can be ported to other operating systems without modifications.
Both versions shares the several features, but the GUI-based version supports only the C language as target. Software packages: Tree2C is provided in three different packages:
The stand-alone version of Tree2C doesn't require the installation, while the GUI version is integrated in VEGA ZZ package and is installed automatically when you run the VEGA ZZ setup. Moreover, the same package includes the command line version (for both 32 and 64 bit Windows versions).
|
| < | Privacy and cookies | > |