| Tree2C Classification tree to code converter |
Main topics:
This program converts the machine learning models, in particular the classification trees, generated by Weka program mainly to C source code but it supports also other programming languages (e.g. C++, Fortran 90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL and VBScript). The resulting code requires no or very limited modifications to be used. The program can recognize several molecular attributes/descriptors (especially those that are calculated by VEGA ZZ and MOPAC 2016) and it can add automatically the code to calculate them. Moreover, Tree2C can generate code also for the domain property check, which is a very useful feature to evaluate the confidence of the classification results. In addition, the program can build the code for different targets to be integrated in a pre-existing program:
The code generated by Tree2C was successfully tested by different language compilers and interpreters as shown in the following table:
| Language | Operating systems | Compiler/Interpreter | Version |
| C | Linux x86 and x64 | gcc | 4.6 and above |
| Windows x86 and x64 | MinGW | 4.6 and above | |
| Windows x86 and x64 | RAD Studio | 10.2.3 | |
| Windows x86 and x64 | TinyC | 0.9.6 | |
| C++ | Linux x86 and x64 | gcc | 4.6 and above |
| Windows x86 and x64 | MinGW | 4.6 and above | |
| Windows x86 and x64 | RAD Studio | 10.2.3 | |
| Fortran 90 | Linux x86 and x64 | gfortran | 4.6 and above |
| Windows x86 and x64 | MinGW | 4.6 and above | |
| Java | Platform independent | Java SE Development Kit | 8 update 12 |
| JavaScript | Platform independent | Node.js | 10.15.3 |
| JScript | Windows x64 | Windows Script Host | 9.0 |
| Lua | Platform independent | Lua | 5.3.5 |
| PHP | Platform independent | PHP | 5.2.5 and above |
| Python | Platform independent | Python | 3.5.1 |
| REBOL | Platform independent | REBOL/View | 2.7.8.3.1 |
| VBScript | Windows x64 | Windows Script Host | 5.8 |
Tree2C supports both Linux (x86 or x64) and Windows (2000/XP/Vista/7/8/8.1/10 x86 or x64) operating systems and requires the HyperDrive runtime library, which is the same used for VEGA ZZ software. Since the program is written in standard C code, it can be ported to other operating systems without modifications.
Tree2C is provided in two different versions:
Both versions shares the several features, but the GUI-based version supports only the C language as target.
Tree2C is provided in three different packages:
Tree2C_X.X.X.zip
This archive includes the command-line version of the program for Linux (x86
and x64) built by gcc and Windows (x86 and x64) built by Mingw32/64 and RAD
Studio 10.2 Tokyo. Moreover, examples and scripts are provided only in this
package and the GUI version of the program is not included.
Vega_ZZ_X.X.X.X_Setup.exe
This setup includes both command-line and GUI versions of Tree2C and
the applicative examples (Prediction of blood-brain
barrier permeation and Prediction of
mutagenicy) ready to use.
Vega_X.X.X.X_Linux_x86-x64-ARM.tar.gz
This archive includes the command-line version of the program for Linux (x86
and x64) built by gcc. Windows and GUI versions are not provided as well as
examples and scripts.
The stand-alone version of Tree2C doesn't require the installation, while the GUI version is integrated in VEGA ZZ package and is installed automatically when you run the VEGA ZZ setup. Moreover, the same package includes the command line version (for both 32 and 64 bit Windows versions).
Linux systems require to set LD_LIBRARY_PATH environment variable to find the hdrive.so dynamic library. You can do it by editing your shell start-up script (e.g. .cshrc for csh or tcsh, .bashrc for GNU bash). For csh/tcsh shell, you must add these lines at the end of the script:
setenv LD_LIBRARY_PATH "<INSTALLATION_PATH> $LD_LIBRARY_PATH" setenv PATH "<INSTALLATION_PATH>:$PATH"
where <INSTALLATION_PATH> is the directory in which Tree2C executable is present.
For sh/bash:
export LD_LIBRARY_PATH="<INSTALLATION_PATH> $LD_LIBRARY_PATH" export PATH="<INSTALLATION_PATH>:$PATH"
For example, if you installed Tree2C for Linux in /usr/local/tree2c
directory, you must set the environment variables (csh/tcsh):
setenv LD_LIBRARY_PATH "$/usr/local/tree2c $LD_LIBRARY_PATH" setenv PATH "/usr/local/tree2c:$PATH"
or (sh/bash):
export VEGADIR="/usr/local/vega" export LD_LIBRARY_PATH="/usr/local/vega $LD_LIBRARY_PATH" export PATH="/usr/local/vega:$PATH"
Finally, you must change the file permissions:
chmod 755 tree2c
Before to use this utility, you must follow these steps in order obtain the code to perform the classification:
preparation of the dataset to teach the learning algorithm;
calculation of the attributes/descriptors;
generation of the classification model by Weka;
generation of the code by Tree2C.
This workflow is especially thought for the classification of molecules, but Tree2C can be used with success also with non-chemical datasets and models.
4.1 Data preparation for machine learning with VEGA ZZ
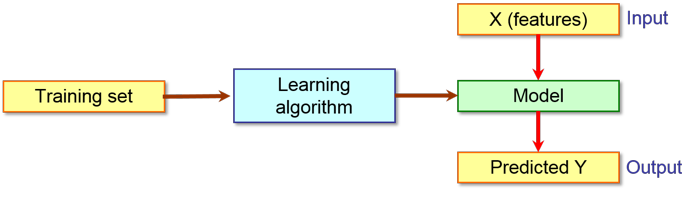
When you want to build a model to classify molecules, a training dataset of examples for each class is required in order to teach the learning algorithm. The so obtained model can be used to predict the belonging class of an unknown molecule as shown in the following scheme:

Obviously, to perform the classification, you
must know the features/attributes of the query molecule and have the right tool
to calculate them.
VEGA ZZ can help you in the first phase of this workflow and, in particular, it
can be used not only to prepare the training set of molecules, but also to
calculate several attributes. Its flexible
database engine can help you in organizing and processing molecules from
different data sources and formats (e.g. IUPAC name, use name, SMILES notation,
InChI notation, 2D and 3D structures in different format, etc.) in order to
obtain homogenous data ready to use for the calculation of the attributes as
shown in the following scheme:
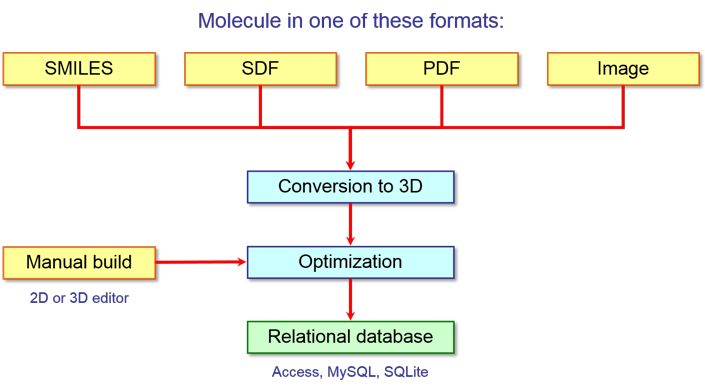
VEGA ZZ supports
different types of databases, but when you have to perform the machine
learning, it is very important the use of relational databases (e.g. Microsoft
Access, MySQL, SQLite) because 1) they can include not only the
molecules structures
but also their attributes and the belonging class; 2) you can
manage the data not only with VEGA ZZ but also with other programs; 3) you can use the
WarpEngine technology to calculate the
descriptors (e.g. semi-empirical ones).
The generic procedure, which you can use to build a training
set with VEGA ZZ, could be:
Create and empty relational database (e.g. in
Access format) by selecting the File
![]() Database
Database
![]() Open item
of main menu or clicking the Open button in the
Database
explorer window.
Open item
of main menu or clicking the Open button in the
Database
explorer window.
Open the empty database in the Database explorer.
Put the molecules into the database as explained here. When you add molecules to a relational database, VEGA ZZ calculates several properties that can be used as attributes for machine learning (look here for a complete list of the properties).
If you want to calculate the Kier-Hall
e-state descriptors, select File
![]() Run script in VEGA ZZ main menu, expand the tree at the
Database level, choose
Count functional groups.c script, and click the Run button.
Select the database to process, in the message box, click No to
store the descriptors in the same database and not a separated CSV file.
Run script in VEGA ZZ main menu, expand the tree at the
Database level, choose
Count functional groups.c script, and click the Run button.
Select the database to process, in the message box, click No to
store the descriptors in the same database and not a separated CSV file.
If you want to calculate semi-empirical descriptors, you can use the MOPAC module of WarpEngine. The descriptors are saved into a CSV file and must be merged manually with the other ones by using a spreadsheet program.
You can add other property columns from other data sources (e.g. Microsoft Excel) to the database by the clipboard: for example, copy a column of Excel and paste it to Edit tab of Database explorer clicking the Paste cols. button. During this operation, you must pay particular attention because 1) the column must have the label in the first row; 2) the number of items to paste must be the same of the molecules in the database; 3) the items must be in the same order of the molecules in the database to avoid misalignment errors. To overcome this last issue, you must remember that the molecules are alphabetically sorted in ascending order using the same algorithm implemented in Excel. So, if you sort the molecules in the same manner in Excel, you overcome this potential problem.
Finally, you can extract the properties/attributes from the database in three different ways: 1) exporting them directly to Microsoft Excel clicking on the molecule list in Database explorer with the right mouse button and selecting Export to Excel, 2) exporting them to a file clicking as above, but selecting Export to file in the context menu; 3) making a query and exporting the data in your preferred front-end program for the database (e.g. Microsoft Access for mdb and accdb files).
4.2 Model generation with Weka
This part of the manual don't want to be exhaustive (this is only a "mini how-to" guide) and more information can be found in Weka manual and tutorials.
Start Weka and choose Explorer as application.
In Process tab, click Open file... and select the ARFF input file if you have it. This file can be prepared saving the data in CSV format from your preferred spreadsheet program (e.g. Microsoft Excel) and import it to Weka. In some cases, the imported file needs to be pre-processed (use the Edit button). More information on ARFF format (Attribute-Relation File Format) is available in Weka manual.
Go to Classify tab and choose the classifier (press Choose button in Classifier box). For example, select RandomForrest in trees.
In the options of the classifier (click on the classifier parameters of Classifier box), set printClassifiers option to True to generate the right output including the trees.
Press Start button to generate the model.
If the model is acceptable, save it by clicking with the right mouse button on Result list and choose Save result buffer. Put the file name adding .txt extension and press Save.
If you run this utility by command prompt without arguments, the program options are shown as here below:
Tree2C V1.0.0 - (c) 2017-2023, Alessandro Pedretti
Usage: Tree2C INPUT_FILE -o[OUTPUT_FILE] -a[DATA_FILE] -i[SCRIPT_DIR]
-l[CLASS_LABELS] -n[MODEL_NAME] -s[LANGUAGE]
-t[TEMPLATE_DIR] -dfhmv
a -> ARFF file to generate the domain check code
d -> Add DLL code (define T2C_DLL to enable compiling the code)
f -> Force to write the code also for the unused attributes
h -> Save all code in the header file
i -> Install the C-script in the specified directory
l -> Class labels (comma separated)
m -> Multi-language support (VEGA ZZ C-script only)
n -> Name of the model (default input file name)
o -> Output file name (default input file)
s -> Target programming language:
C (default), C++, Fortran90, Java, JavaScript, JScript, Lua,
PHP, Python, REBOL, VBScript
t -> VEGA template directory (usually autodetected)
v -> Code compatible with VEGA ZZ C-script
Examples:
Tree2C weka_tree.txt
Tree2C weka_tree.txt -s Python
Tree2C weka_tree.txt -l "No,Yes"
Tree2C weka_tree.txt -o prediction.c -a weka_input.arff -v
All parameters are optional with the exception of the the input file (INPUT_FILE), which includes the decision tree model generated as explained in the previous section. The meaning of the other parameters is summarized in the following table:
| Option | Argument | Description |
| -a | DATA_FILE | If you specify the ARFF file used to create the model, additional code is generated to check if the calculated attributes are included in the same domain as those used to build the model. |
| -d | - | Add the code to compile the model as dynamic link library (DLL) for Windows OSs. Define T2C_DLL if you want to obtain a DLL, otherwise the resulting object will be the same as without -d option. This option is available only for the C target language. |
| -f | - | Usually Tree2C doesn't consider the attributes not used by Weka in the tree even if they appear in the header of the model or ARFF files. By this switch, you can force the code generation for all attributes. This feature is useful when you want to use together more than one model sharing the same set of attributes. |
| -h | - | The code is merged in the header file (.h) without to create the C file (.c). |
| -i | SCRIPT_DIR | Install the code as C-script the specified script directory (see -v option). This feature is useful only if VEGA ZZ is installed. |
| -l | CLASS_LABELS | Usually Tree2C uses a progressive number to indicate each class for the prediction (e.g. 0, 1 ... n), but you can change this behaviour specifying a label for each class. Each label must be comma separated and included between quotes (e.g. "Inactive,Active" respectively for 0 and 1 classes). |
| -m | - | Enable the language localization of the C-script (see -v option). |
| -n | MODEL_NAME | Name of the model. By default, it is the input file name without path and extension. |
| -o | OUTPUT_FILE | Name of the output file(s). By default, the input file name is used for both code and header files. |
| -s | LANGUAGE | Target programming language used to generate the source code. Actually, the keywords for the supported languages are: C, C++, Fortran90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL, and VBScript. This option is case-insensitive. |
| -t | TEMPLATE_DIR | Full path of the VEGA template directory needed to calculate the some molecular descriptors. |
| -v | - | Generate the code as VEGA ZZ C-script, including the code to calculate the known molecular properties. When you run the resulting script, if the VEGA workspace is not empty and there is a molecule, the classification is performed for the current molecule, otherwise a file requester is shown to select an input database. In this second case, the classification is performed for all molecules of the database. The output is a CSV file whose name can be specified through a file requester. |
*All options are case-insensitive.
When you generate the code as VEGA ZZ C-script (-v option), the attribute names are analyzed and if are calculable by VEGA ZZ, the right code is automatically added to the output, otherwise a warning message is shown. In this case, you have to complete the code.
Here are some examples to clarify the use of Tree2C:
Here, the typical Tree2C output of a generic run is shown:
tree2c "random tree.txt" * Loading the model Target programming language..: C Original model name..........: RANDOM_TREE Model name for the code......: RANDOM_TREE Number of attributes.........: 58 Number of unused attributes..: 31 Unused attributes............: Surface Charge FG_Br FG_CHO FG_CN3 FG_CNH FG_CNR Number of trees..............: 1 Number of output values......: 2 Class values.................: FALSE TRUE Class labels.................: None * Saving the C code * Done
4.4 Graphic user interface
To use the GUI version of Tree2C, you must start VEGA ZZ
and select File
![]() Run script
in the main menu. Hence, you must find
Development tools
Run script
in the main menu. Hence, you must find
Development tools
![]() Decision tree to C converter.vll in the script tree and finally you must double
click on it. Although the program is managed as a VEGA ZZ
C-script, you cannot edit the source code because it was built as VEGA Link
Library (VLL), which is in binary format. As for the standard C-scripts, you can
show the help clicking the > symbol on the right side of the Select the
script ... window.
Decision tree to C converter.vll in the script tree and finally you must double
click on it. Although the program is managed as a VEGA ZZ
C-script, you cannot edit the source code because it was built as VEGA Link
Library (VLL), which is in binary format. As for the standard C-scripts, you can
show the help clicking the > symbol on the right side of the Select the
script ... window.
The features of this Tree2C version are the same of the command line version but are accessible through a nice graphic user interface. To build the C code (the only language supported by this version), you must:
As explained above, the input file required by Tree2C is the output
generated by Weka when you run a tree-based classifier (random tree,
random forest, etc.), but since is a text file, you can generate it in
easy way by other programs. This paragraph doesn't want to be an exhaustive
guide on the Weka output format, but shows only the most important topics aimed
to build a file compatible with Tree2C.
The file includes three parts: 1) a header with the information on the
attributes and the learning approach (Run information section); 2) one or
more decision trees (Classifier model section); 3) a footer with the
statistical data (Summary section). Tree2C requires only the first
two sections of the file.
Not all tags of this section are needed by Tree2C and, in particular, only the first two must be always present: Scheme and Attributes.
Scheme: weka.classifiers.trees.RandomTree -K 0 -M 1.0 -V 0.001 -S 1and the minimal value of Scheme for Tree2C is:
Scheme: weka.classifiers.trees
Attributes: 5 Angles Atoms Dipole HbAcc HbDonSince Tree2C can find the attributes directly from the trees, you cannot specify their list as shown below:
Attributes: 5 [list of attributes omitted]
The Run information label cannot be present because Tree2C assumes by default that the first section found in the file is just this one.
4.5.2 Classifier model section
The Classifier model label marks the section in which one ore more decision trees are reported. This label is optional for Tree2C because it searches directly for the RandomTree label, which denotes the beginning of each tree. The tree is drawn from left to right and the splitting nodes are not indicated but are placed virtually in the middle of the segment built by multiple pipe characters ( | ). At the end of two branches of the fork there is a leaf represented by an attribute and a condition (respectively the true and false conditions for each leaf pair) as shown below:
Attribute1 < Value1 | | Other branches | Attribute1 >= Value1 | | Other branches
The previous example shows the comparison of an attribute (Attribute1) with a threshold value (Value1) through a pair of operators (less than and equal or greater than) for both true and false conditions. Just for an exemplification, the corresponding pseudo-code is:
if Attribute1 < Value1 then
True condition
else
False condition
If you have to insert a branch with a Boolean attribute, you must use a different representation as shown below:
Attribute2 = yes | | Other branches | Attribute2 = no | | Other branches
and the corresponding pseudo code is:
if Attribute2 is true then
True condition
else
False condition
When a leaf must return the class and you don't have to continue the tree with other branches, you can indicate the class after the condition as shown in the following example:
Attribute3 < Value3 : 1 Attribute3 >= Value3 : 0
where 0 and 1 are the class IDs. Translating the node to pseudo-code:
if Attribute3 > Value3 then
return 1
else
return 0
Weka adds statistical data (two integer numbers separated by a slash) to this kind of leaf, which is however ignored by Tree2C:
Attribute3 < Value3 : 1 (TotInst/MissClassInst) Attribute3 >= Value3 : 0 (TotInst/MissClassInst)
In particular, the first number is the total number of instances (TotInst, weight of instances) reaching the leaf and the second number is the number (weight) of those instances that are misclassified (MIssClassInst).
All constants, data structures and functions are named according to the rule in which each object has T2C_MODEL_NAME_ prefix, where MODEL_NAME is obtained automatically from the Weka file name capitalizing it and replacing the spaces with underscores characters ("_") or is specified by the user with -n option.
The constants are defined in the header file or in the C file if you have selected an output without header file.
/**** Output values ****/ #define T2C_RANDOM_TREE_FALSE 0 #define T2C_RANDOM_TREE_TRUE 1
/**** Values of the attributes ****/ #define T2C_RANDOM_TREE_FG_CON2_NO 0 #define T2C_RANDOM_TREE_FG_CON2_YES 1
/**** MOPAC keywords ****/ #define T2C_MOPAC_KEYS "PM7 GEO-OK MMOK 1SCF SUPER" /**** MOPAC properties ****/ #define T2C_MOPAC_CORE_CORE_REPULSION 0 #define T2C_MOPAC_COSMO_AREA 1 #define T2C_MOPAC_COSMO_VOLUME 2 #define T2C_MOPAC_DE_TOTAL 3 #define T2C_MOPAC_DN_TOTAL 4 #define T2C_MOPAC_DIPOLE 5 #define T2C_MOPAC_ELECTRONIC_ENERGY 6 #define T2C_MOPAC_HEAT_OF_FORMATION 7 #define T2C_MOPAC_HOMO_ENERGY 8 #define T2C_MOPAC_IONIZATION_POTENTIAL 9 #define T2C_MOPAC_LUMO_ENERGY 10 #define T2C_MOPAC_MULLIKEN_ELECTRONEGATIVITY 11 #define T2C_MOPAC_NO_OF_FILLED_LEVELS 12 #define T2C_MOPAC_PARR_POPLE_ABSOLUTE_HARDNESS 13 #define T2C_MOPAC_PIS_TOTAL 14 #define T2C_MOPAC_SCHUURMANN_MO_SHIFT_ALPHA 15 #define T2C_MOPAC_TOTAL_ENERGY 16 #define T2C_MOPAC_NUM_OF_PROPERTIES 17
| Constant | Description |
| T2C_ABWWIDTH | Width of the abort window. |
| T2C_ABWHEIGHT | Height of the abort window. |
| T2C_BTWIDTH | Width of the abort button. |
| T2C_BTHEIGHT | Height of the abort button. |
Example:
/**** Window parameters ****/ #define T2C_ABWWIDTH 300 /* Abort window width */ #define T2C_ABWHEIGHT 104 /* Abort window height */ #define T2C_BTWIDTH 89 /* Button width */ #define T2C_BTHEIGHT 25 /* Button height */
The only data type defined in the source code is that is used as input for both T2C_MODEL_NAME_Classify() and T2C_MODEL_NAME_DomCheck() functions. It is named T2C_MODEL_NAME_INPUT and in its structure, the Boolean and discrete attributes/parameters are defined respectively as integer numbers (int type) and the other ones as single precision floating point numbers (float type).
Example:
/**** Data types ****/ typedef struct { float Angles; float Atoms; float Bonds; float ChiralAtms; float Dipole; float EzBnds; int FG_CON2; int FG_COOH; int FG_COOR; int FG_F; int FG_PhOH; float FlexTorsions; float Gyrrad; float HbAcc; float HbDon; float HeavyAtoms; float Lipole; float Mass; float Ovality; float Psa; float Rings; float Sas; float Sav; float Sdiam; float Torsions; float Vdiam; float VirtualLogP; } T2C_RANDOM_TREE_INPUT;
Tree2C declares several shared global variables for the C-scripts as shown in the following table:
| Name | Type | Header | Description |
| Errors | HD_ULONG | hdtypes.h | Number of errors occurred during the classification. |
| FH | FILE * | stdio.h | Pointer to the file handle used to write the output CSV file. |
| hAbort | GAZ_WINDOW | graphappz.h | Handle of the abort window. |
| hDb | HD_STRING | hdtypes.h | String variable, which contains the handle of the library/database to process. |
| hMopac | HD_PROC | hdprocess.h | Process handle for MOPAC calculations. |
| hThread | HD_THREAD | hdtypes.h | Handle of the calculation thread (used only when you have to process a library of molecules). |
| LBL_Abort | GAZ_LABEL | graphappz.h | Handle of the label gadget showing the progress messages in the abort window. |
| Mols | HD_LONG | hdtypes.h | Number of the molecules included in the library to which to predict the belonging class. |
| MopacProp | float | - | Vector with T2C_MOPAC_NUM_OF_PROPERTIES size used to store the semi-empirical descriptors calculated by MOPAC. |
| Running | HD_BOOL | hdtypes.h | This variable signals to the working process to abort the calculation when it is set to 0. |
Here you can find the description of each C function generated by Tree2C.
| Parameters: | ||
| Input | Attributes/descriptors needed for the classification that must be pre-calculated. | |
Return values:
This function returns the class as integer number (usually 0 and 1) according to
the given input data.
| Parameters: | ||
| Input | Attributes/descriptors needed for the classification that must be pre-calculated. | |
Return values:
This function returns the number of
domain violations, which is a measure of the reliability of data: the higher
number of violations, the lower the reliability of the prediction. It ranges
from 0 (= no violations) to the total number of the attributes included in
the classification model (= lowest reliability of data).
Here is the list of the specific function for C-scripts. Some of them uses GraphAppZ and HyperDrive data types that are defined respectively in graphappz.h and hdtype.h header files that are stored in ...\VEGA ZZ\Tcc\include\vega and ...\VEGA ZZ\Tcc\include\hyperdrive directory. If you installed the 64 bit version of VEGA ZZ, the Tcc directory is renamed to Tcc64.
| Parameters: | ||
| b | Handle of the button generating the event. | |
Return values:
This function doesn't return any value.
| Parameters: | ||
| Arg | Pointer to user data used by the thread that, in this case, is set to NULL. | |
Return values:
This function returns a non-zero value if an error occurs.
| Parameters: | ||
| Err | Pointer to the C string with the error message. | |
Return values:
This function doesn't return any value.
| Parameters: | ||
| hProc | Handle of the process created with HD_ProcNew() of HyperDrive library. | |
| MopacKeys | MOPAC keywords to control the calculation (see T2C_MOPAC_KEYS constants). | |
Return values:
Return values can be 1 if no error occurs, or 0 if the function fails.
| Parameters: | ||
| Class |
Pointer to the integer in which the function returns the predicted class. |
|
| Violations | Pointer to the integer in which the function returns the number of the domain violations. | |
| Atm | Pointer of the first element of the atom list according to the VEGA convention. | |
| TotAtm | Total number of the atoms of the molecule to which you want to perform the classification. | |
Return values:
Return values can be 1 if no error occurs, or 0 if the function fails.
| Parameters: | ||
| Att |
Attribute/descriptor name. |
|
Return values:
This function returns the calculated value as floating point number.
When you select C++ as output, Tree2C generates two files: the header
and the code files. The former includes the definition of the class which is
named according to T2CPP_MODEL_NAME rule and the latter includes the code
of the methods. Tree2C is
unable to generate the C++ code to calculate the attributes/parameters when you
specify
the -v option.
The constants are defined in the header file as for the C output.
The model class includes properties and methods to perform the classification in easy way and is defined as in the following example:
/**** Class definition ****/ class T2CPP_RANDOM_TREE { public:
The properties are defined in the header file as shown in the following example:
/**** Properties ****/ float Angles; float Atoms; int FG_CON2;
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them.
Before to use the class, you must create the related object:
T2CPP_RANDOM_TREE Model;
then, you must set the properties:
Model.Angles = 325.0f; Model.Atoms = 176.0f; Model.FG_CON2 = 0;
finally, you must call the methods:
printf("Domain violations: %d\n", Model.DomCheck());
printf("Predicted class: %d\n", Model.Classify());
The resources of Model object are automatically freed exiting by the function and/or the compound. The object can be also created dynamically by new command:
T2CPP_RANDOM_TREE * Model = new T2CPP_RANDOM_TREE();
but since new returns the pointer to the object, the syntax required to address properties and methods is different and, in particular, for the properties:
Model -> Angles = 325.0f; Model -> Atoms = 176.0f; Model -> FG_CON2 = 0;
and for the methods:
printf("Domain violations: %d\n", Model -> DomCheck());
printf("Predicted class: %d\n", Model -> Classify());
The resources of the objects created by new aren't not automatically released and, therefore, you must free them by delete command:
delete Model;
7. Fortran 90 code description
When you select Fortran90 as output, the class/module is named according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the Fortran 90 code to calculate the attributes/parameters when you specify the -v option.
The model module includes constants, properties and methods to perform the classification in easy way and is defined as in the following example:
!**** Classification module ****/ module random_tree implicit none
The constants have T2F90_MODEL_NAME_ prefix and are defined inside the module as parameters:
!**** Output values **** integer, parameter :: T2F90_RANDOM_TREE_FALSE = 0 integer, parameter :: T2F90_RANDOM_TREE_TRUE = 1
!**** Values of the attributes **** integer, parameter :: T2F90_RANDOM_TREE_FG_CON2_NO = 0 integer, parameter :: T2F90_RANDOM_TREE_FG_CON2_YES = 1
The properties are declared inside a type definition whose name is T2F90_MODEL_NAME_Input as shown in the following example:
type :: T2F90_RANDOM_TREE_Input
!**** Properties ****
real :: Angles
real :: Atoms
integer :: FG_CON2
The type definition is closed by the declaration of the methods to perform the classification (Classify) and the domain check (DomCheck):
contains
procedure, pass(this) :: Classify
procedure, pass(this) :: DomCheck
end type T2F90_RANDOM_TREE_Input
Tree2C generates the code for two methods Classify and, optionally when you specify the ARFF file with -a option, DomCheck. Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, after the program declaration, you must include the module code:
program random_tree_test use random_tree implicit none
before to use the class, you must create the related object:
type(T2F90_RANDOM_TREE_Input) :: Model
then, you must set the properties:
Model%Angles = 325 Model%Atoms = 176 Model%FG_CON2 = 0
and now you can call the methods:
print *, " Domain violations: ", Model%DomCheck() print *, " Predicted class: ", Model%Classify()
finally, you must remember to add the following line to end the program:
end program random_tree_test
When you select Java as output, the class is named (in lower case) according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the Java code to calculate the attributes/parameters when you specify the -v option.
The model class includes constants, properties and methods to perform the classification in easy way and is defined as in the following example:
/**** Model class ****/
class random_tree {
The constants have C_ prefix and are defined inside the class as static final properties (constants):
/**** Output values ****/ static final int C_FALSE = 0; static final int C_TRUE = 1;
/**** Values of the attributes ****/ static final int C_FG_CON2_NO = 0; static final int C_FG_CON2_YES = 1;
The properties are declared inside the class as shown in the following example:
/**** Attributes ****/ float Angles; float Atoms; int FG_CON2;
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, after the program and main class declaration, you create the object of the class:
public class random_tree_test
{
public static void main(String[] args)
{
random_tree Model = new random_tree();
then, you must set the properties:
Model.Angles = 325.0f; Model.Atoms = 176.0f; Model.FG_CON2 = 0;
and now you can call the methods:
System.out.printf(" Domain violations: %d\n", Model.DomCheck());
System.out.printf(" Predicted class: %d\n", Model.Classify());
finally, you must remember to add the following lines to end the program:
} }
9. JavaScript code description
When you select JavaScript as output, the class is named (in lower case) according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the JavaScript code to calculate the attributes/parameters when you specify the -v option.
The model class includes constants, properties and methods to perform the classification in easy way and is defined as in the following example:
/**** Model class ****/
class random_tree {
The constants (this is an improper term, because in JavaScript the constant type doesn't exist) have C_ prefix and are defined inside the constructor of the class:
/**** Constructor ****/
constructor() {
There are two type of constants:
/**** Output values ****/
this.C_FALSE = 0;
this.C_TRUE = 1;
/**** Values of the attributes ****/
this.C_FG_CON2_NO = 0;
this.C_FG_CON2_YES = 1;
The properties are declared inside the class constructor and are initialized to zero as shown in the following example:
/**** Attributes ****/
this.Angles = 0.0;
this.Atoms = 0.0;
this.FG_CON2 = 0;
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, you must include the code of the classification class. The following example is based on Node.js but the syntax can differ on the basis of the JavaScript interpreter:
const random_tree = require('./random_tree.js');
before to use the class, you must create the related object:
const Model = new random_tree();
and now you must set the properties:
Model.Angles = 325.0; Model.Atoms = 176.0; Model.FG_CON2 = 0;
finally, you can call the methods:
console.log(" Domain violations: %d", Model.DomCheck());
console.log(" Predicted class: %d", Model.Classify());
JScript is Microsoft's dialect of ECMAScript standard and although shares
some programming and syntax paradigms with JavaScript, it must not confused with
JavaScript. JScript doesn't support class programming, but it is possible to
implement pseudo-classes through functions.
When you select JScript as output, a pseudo-class is created as a
function
named (in lower case) according to the name of the model in Weka file or
by -n option. Tree2C is
unable to generate the JScript code to calculate the attributes/parameters when you
specify
the -v option.
The model class includes constants, properties and methods to perform the classification in easy way and is defined as in the following example:
/**** Model pseudo-class ****/
function random_tree() {
As for JavaScript, the constants are not supported by JScript, but Tree2C creates the code declaring and initializing variables according to the rule in which each variable is preceded by the C_ prefix. They are defined at the beginning of the main function and there are two type of constants:
/**** Output values ****/
this.C_FALSE = 0;
this.C_TRUE = 1;
/**** Values of the attributes ****/
this.C_FG_CON2_NO = 0;
this.C_FG_CON2_YES = 1;
The properties are declared inside the main function and are initialized to zero as shown in the following example:
/**** Attributes ****/
this.Angles = 0.0;
this.Atoms = 0.0;
this.FG_CON2 = 0;
Tree2C generates the code for two functions inside the main function, namely Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both functions can be managed as methods and don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two functions, click here.
As first step, you must include the code of the classification function.
<job> <script language="JScript" src="random_tree_jscript.js"/> <script language="JScript">
before to use the external function, you must create the related object:
var Model = new random_tree();
then, you must set the properties:
Model.Angles = 325.0;
Model.Atoms = 176.0;
Model.FG_CON2 = 0;
finally, you can call the methods:
WSH.echo("Domain violations:", Model.DomCheck(),
"Predicted class: ", Model.Classify());
finally, you must remember to add the following lines to end the script:
</script> </job>
When you select Lua as output, Tree2C generates the the class code for you, which is a little bit tricky if you want to write it by yourself. As usual, the class is named (in lower case) according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the Lua code to calculate the attributes/parameters when you specify the -v option.
As explained above, all code to declare the class is automatically generated by Tree2C as shown in the following example:
---- Model class ----
random_tree = {}
random_tree.__index = random_tree
Also for Lua, the constants are not supported and are replaced by standard properties which, however, are not write-protected as the constants. This kind of properties, have C_ prefix and are defined inside the constructor of the class:
function random_tree:New()
local Acnt = {}
setmetatable(Acnt, random_tree)
There are two type of constants:
---- Output values ---- self.C_FALSE = 0 self.C_TRUE = 1
/**** Values of the attributes ****/
this.C_FG_CON2_NO = 0;
this.C_FG_CON2_YES = 1;
The properties are declared inside the class constructor and are initialized to zero as shown in the following example:
---- Properties ---- self.Angles = 0.0 self.Atoms = 0.0 self.FG_CON2 = 0
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, you must include the code of the classification class:
require 'random_tree'
before to use the class, you must create the related object:
Model = random_tree:New()
then, you must set the properties:
Model.Angles = 325 Model.Atoms = 176 Model.FG_CON2 = 0
finally, you can call the methods:
print(" Domain violations: " .. Model:DomCheck())
print(" Predicted class: " .. Model:Classify())
When you select PHP as output, the class is named (in upper case) according to the name of the model in Weka file or by -n option preceded by T2PHP_ prefix. Tree2C is unable to generate the PHP code to calculate the attributes/parameters when you specify the -v option.
The model class includes constants, properties and methods to perform the classification in easy way and is defined as in the following example::
/**** Model class ****/
class T2PHP_RANDOM_TREE {
There are two type of constants, which are declared inside the class and have C_ prefix:
/**** Output values ****/ const C_FALSE = 0; const C_TRUE = 1;
/**** Values of the attributes ****/ const C_FG_CON2_NO = 0; const C_FG_CON2_YES = 1;
The properties are declared inside the class constructor and are initialized to zero as shown in the following example:
/**** Properties ****/ public $Angles = 0.0; public $Atoms = 0.0; public $FG_CON2 = 0;
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, you must include the code of the classification class:
<?php require 'random_tree.php';
before to use the class, you must create the related object:
$Model = new T2PHP_RANDOM_TREE();
then, you must set the properties:
$Model -> Angles = 325; $Model -> Atoms = 176; $Model -> FG_CON2 = 0;
and now you can call the methods:
echo " Domain violations: " . $Model -> DomCheck() . "\n"; echo " Predicted class: " . $Model -> Classify() . "\n";
finally, you must remember to add the following line to end the script:
?>
The code generated by Tree2C is compatible only with Python 3 and Python 2.7 is not currently supported. More in detail, as for the C code, the class is named as T2PY_MODEL_NAME. Although VEGA ZZ supports Python for scripting, at this time, Tree2C is unable to generate the code to calculate the attributes/parameters when you use the -v option.
The model class includes the properties/attributes and the methods to perform the classification and the domain check in easy way and is defined as in the following example:
#**** Model class **** class T2PY_RANDOM_TREE:
Unlike the C language, which supports constants values through the macro pre-processor, Python doesn't have this feature and hence the constants are defined as standard properties inside the class of the classifier.
#**** Output values **** C_FALSE = 0 C_TRUE = 1
#**** Values of the attributes **** C_FG_CON2_NO = 0 C_FG_CON2_YES = 1
Although Python doesn't require to declare the properties, it was preferred initialize all attributes to 0 in order to provide a list of those that are effectively used in the model, making easier the code writing to pass the values. Example:
#**** Properties **** Angles = 0.0 Atoms = 0.0 FG_CON2 = 0
Tree2C generates the code for two methods Classify() and, optionally when you specify the ARFF file with -a option, DomCheck(). For more information, see the C++ version.
First of all, you must include the code of the classification class:
import random_tree
before to use the class, you must create the related object:
Model = random_tree.T2PY_RANDOM_TREE()
then, you must set the properties:
Model.Angles = 325 Model.Atoms = 176 Model.FG_CON2 = 0
finally, you can call the methods:
print(" Domain violations:", Model.DomCheck())
print(" Predicted class: ", Model.Classify())
When you select REBOL as output, the class is named (in lower case) according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the REBOL code to calculate the attributes/parameters when you specify the -v option.
The model class includes constants, properties and methods to perform the classification in easy way and is defined as in the following example::
REBOL [ Title: "random_tree" File: %random_tree.r ] ;**** Model class **** random_tree: make object! [
REBOL doesn't support constants, which therefore are implemented as normal class properties. There are two type of constants, which are declared inside the class and have C_ prefix:
;**** Output values **** C_FALSE: 0 C_TRUE: 1
;**** Values of the attributes **** C_FG_CON2_NO: 0 C_FG_CON2_YES: 1
The properties are declared inside the class and are initialized to zero as shown in the following example:
;**** Properties **** Angles: 0.0 Atoms: 0.0 FG_CON2: 0
Tree2C generates the code for two methods Classify and, optionally when you specify the ARFF file with -a option, DomCheck. Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
First of all, you must include the code of the classification class:
REBOL [ Title: "random_tree_test" File: %random_tree_test.r ] do %random_tree.r
the object don't need to be created and you can set directly the properties:
random_tree/Angles: 325.0 random_tree/Atoms: 176.0 random_tree/FG_CON2: 0
and now you can call the methods:
print [" Domain violations: " random_tree/DomCheck] print [" Predicted class: " random_tree/Classify "]
When you select VBScript as output, the class is named (in lower case) according to the name of the model in Weka file or by -n option. Tree2C is unable to generate the VBScript code to calculate the attributes/parameters when you specify the -v option.
There are two type of constants, which are declared with T2VBS_MODEL_NAME_ prefix:
'**** Output values **** const T2VBS_RANDOM_TREE_FALSE = 0 const T2VBS_RANDOM_TREE_TRUE = 1
'**** Values of the attributes **** const T2VBS_RANDOM_TREE_FG_CON2_NO = 0 const T2VBS_RANDOM_TREE_FG_CON2_YES = 1
The model class includes properties and methods to perform the classification in easy way and is defined as in the following example::
'**** Model class **** Class random_tree
The properties are declared inside the class and to avoid conflicts with VBS keywords, the Attr suffix is added to the name as shown in the following example:
'**** Attributes **** Public AnglesAttr Public AtomsAttr Public FG_CON2Attr
Tree2C generates the code for two methods Classify and, optionally when you specify the ARFF file with -a option, DomCheck. Both methods don't have arguments because the data is taken from the attribute properties that must be set before to call them. For more information on these two methods, click here.
As first step, you must include the code of the classification function.
<job> <script language="VBScript" src="random_tree.vbs"/> <script language="VBScript">
before to use the external function, you must create the related object:
Dim Model
Set Model = New random_tree
then, you must set the properties:
Model.AnglesAttr = 325.0
Model.AtomsAttr = 176.0
Model.FG_CON2Attr = 0
finally, you can call the methods:
WScript.Echo "Domain violations:", Model.DomCheck(), vbNewLine, _
"Predicted class: ", Model.Classify()
finally, you must remember to add the following lines to end the script:
</script> </job>
16.1 Prediction of blood-brain barrier permeation
This is an example of a C-script generated automatically by Tree2C and
performs the classification of molecules between permeants and non-permeants of
blood-brain barrier (BBB) through a decision tree. Since the attributes are
calculated by VEGA ZZ, no additional code was written manually. This script is
included in VEGA ZZ package and you can run it selecting File
![]() Run script
in VEGA ZZ main menu and double clicking BBB permeation predictor.c in
ADMET.
Run script
in VEGA ZZ main menu and double clicking BBB permeation predictor.c in
ADMET.
If a molecule is present in the current workspace, a single classification is
performed, otherwise a file requester is shown to select an input database. In
this case, the classification is performed for all molecules of the database and
the results are saved to a CSV file.
Since the training set used in the learning phase to build the model includes
molecules is in neutral form, also the molecules for which you want to predict the
BBB permeation must be in this form.
16.1.2 About the decision tree model
To derive the model, the Li's dataset (J. Chem. Inf. Model., 2005, 45, 1376-1384) was used as learning set with Weka 3.8 software. All molecules were converted from SMILES to 3D by VEGA ZZ and optimized by MOPAC 2016 (PM7 PRECISE GEO-OK SUPER keywords), keeping them in neutral form. 129 properties/attributes were calculated by both VEGA ZZ and MOPAC 2016. The most significant attributes were selected according to the BestFirst search algorithm (direction = Forward; lookupCacheSize = 1; searchTermination = 5) and the WrapperSubsetEval attribute evaluator (classifier = RandomForest with default settings; doNotCheckCapabilities = False; evaluationMeasure = accuracy, RMSE; folds = 5; seed = 1; threshold = 0.01) as implemented in Weka. In this way, only 9 attributes were kept, namely:
| Attribute | Description |
| Bonds | Number of bonds |
| Charge | Total charge |
| HeavyAtoms | Number of heavy atoms |
| Mass | Molecule mass |
| Vdiam | Volume diameter |
| VirtualLogP | Molecular lipophilicity cacluated as Log P according to Testa's method |
| FG_aaNH | Kier-Hall E-state descriptor |
| FG_sCH3 | Kier-Hall E-state descriptor |
| FG_sssN | Kier-Hall E-state descriptor |
Charge attribute appears in the list because the learning set includes
quaternary ammonic molecules that were not neutralized with a counter ion. All
electronic descriptor calculated by MOPAC 2016 with SUPER keyword were
considered not meaningful by the selection algorithm to be considered in the
next phase.
The final model was obtained by Random Forest machine learning algorithm
implemented in Weka with default parameters (bagging with 100 iterations and
base learner) and performing a 10 fold cross-validation. The results are
summarized below, where class 0 and 1 indicate non-permenat and permeant
molecules:
=== Stratified cross-validation ===
=== Summary ===
Correctly Classified Instances 346 83.3735 %
Incorrectly Classified Instances 69 16.6265 %
Kappa statistic 0.6179
Mean absolute error 0.2655
Root mean squared error 0.3647
Relative absolute error 59.5558 %
Root relative squared error 77.2667 %
Total Number of Instances 415
=== Detailed Accuracy By Class ===
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.705 0.101 0.778 0.705 0.740 0.620 0.865 0.787 0
0.899 0.295 0.858 0.899 0.878 0.620 0.865 0.912 1
Weighted Avg. 0.834 0.230 0.831 0.834 0.832 0.620 0.865 0.870
=== Confusion Matrix ===
a b <-- classified as
98 41 | a = 0
28 248 | b = 1
The overall accuracy of the model after cross validation is 83.37%, value which is comparable to that of 83.7% obtained by support vector machine (SVM) with recursive feature elimination (RFE) as published by Li et al. Also the Matthews correlation coefficient (MCC) is quite similar for both models (0.620 vs. 0.645), but the Weka model uses only 9 descriptors instead of 35 of the SVM+RFE model and doesn't include QM properties, reducing dramatically the time required to calculate the parameters. Finally, both models are based on Kier-Hall E-state descriptors but for the random forest model, only 3 descriptors are taken into account instead of 17 of the SVM+RFE model. This is a further advantage in reducing the time for the prediction.
As the previous example, this is a C-script, which was generated automatically by Tree2C with the aim to classy between mutagen and non-mutagen molecules through a decision model obtained by machine learning. Also in this case, all attributes were calculated by VEGA ZZ and so no additional code was written manually.
Since this script shares the same base code of the previous example, you can
use it in the same way and, in particular, if a molecule is present in the
current workspace, a single classification is performed, otherwise a file
requester is shown to select an input database and all molecules of the database
are classified saving the result to a CSV file.
Since the training set used in the learning phase to build the model includes
molecules in neutral form, also the molecules for which you want to predict the
mutagenicity must be in this form.
To derive the model, the Bursi's dataset (J. Med. Chem., 2005, 48, 312-320) was used as learning set with Weka 3.8 software. All molecules were converted from SMILES to 3D by VEGA ZZ and optimized by MOPAC 2016 (PM7 PRECISE GEO-OK SUPER keywords) keeping them in neutral form. 129 properties/attributes were calculated by both VEGA ZZ and MOPAC 2016. The most significant attributes were selected according to the BestFirst search algorithm (direction = Forward; lookupCacheSize = 1; searchTermination = 5) and the WrapperSubsetEval attribute evaluator (classifier = RandomForest with default settings; doNotCheckCapabilities = False; evaluationMeasure = accuracy, RMSE; folds = 5; seed = 1; threshold = 0.01) as implemented in Weka. In this way, 24 attributes were kept, namely:
| Attribute | Program | Description |
| EzBnds | VEGA ZZ | Number of asymmetric double bonds |
| HbAcc | VEGA ZZ | Number of H-bond acceptors |
| HbDon | VEGA ZZ | Number of H-bond donors |
| Impropers | VEGA ZZ | Number of improper (out-of-plane) angles |
| Psa | VEGA ZZ | Polar surface area |
| Rings | VEGA ZZ | Number of rings |
| Torsions | VEGA ZZ | Number of dihedrals (torsion angles) |
| HEAT_OF_FORMATION | MOPAC | Heat of formation |
| ELECTRONIC_ENERGY | MOPAC | Electronic energy |
| LUMO_ENERGY | MOPAC | Lumo energy |
| MOLECULAR_WEIGHT | MOPAC | Mass |
| CHARGE_ON_SYSTEM | MOPAC | Total charge |
| PARR_&_POPLE_ABSOLUTE_HARDNESS | MOPAC | Parr & Pople absolute hardness |
| FG_aaCH | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_aaO | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_aasC | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ddssS | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_sCl | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ssCH2 | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ssGeH2 | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ssNH | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ssO | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_sssCH | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_sssN | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_ssssC | VEGA ZZ | Kier-Hall E-state descriptor |
| FG_tCH | VEGA ZZ | Kier-Hall E-state descriptor |
As in the previous example, CHARGE_ON_SYSTEM attribute appears in the list because the learning set includes
quaternary ammonic molecules that were not neutralized with a counter ion. Some
MOPAC attributes were kept during the variable selection procedure and
therefore this script requires MOPAC 2016 to perform the classification.
The final model was obtained by Random Forest machine learning algorithm
implemented in Weka with default parameters (bagging with 100 iterations and
base learner) and performing a 10 fold cross-validation. The results are
summarized here, where class 0 and 1 indicate respectively non-mutagen and
mutagen substances:
=== Stratified cross-validation ===
=== Summary ===
Correctly Classified Instances 3575 82.4303 %
Incorrectly Classified Instances 762 17.5697 %
Kappa statistic 0.6442
Mean absolute error 0.2811
Root mean squared error 0.3615
Relative absolute error 56.8678 %
Root relative squared error 72.7173 %
Total Number of Instances 4337
=== Detailed Accuracy By Class ===
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.798 0.155 0.806 0.798 0.802 0.644 0.896 0.867 0
0.845 0.202 0.838 0.845 0.842 0.644 0.896 0.910 1
Weighted Avg. 0.824 0.181 0.824 0.824 0.824 0.644 0.896 0.891
=== Confusion Matrix ===
a b <-- classified as
1545 391 | a = 0
371 2030 | b = 1
The overall accuracy of Weka model is 82.43%, which is in accordance with the results obtained by Bursi et al. and, in particular, they found a mean accuracy of 82.50%.
All trademarks and software directly or indirectly referred in this document, are copyrighted from legal owners. Tree2C is a freeware program and can be spread through Internet, BBS, CD-ROM and other electronic formats. The Author of this program accepts no responsibility for hardware/software damages resulting from the use of this package. No warranty is made about the software or its performance. Use and copying of this software and the preparation of derivative works based on this software are permitted, so long as the following conditions are met:
The copyright notice and this entire notice are included
intact and prominently carried on all copies and supporting documentation.
No fees or compensation are charged for use, copies, or
access to this software. You may charge a nominal distribution fee for the
physical act of transferring a copy, but you may not charge for the program
itself.
Any work distributed or published that in whole or in part contains or is a derivative of this software or any part thereof is subject to the terms of this agreement. The aggregation of another unrelated program with this software or its derivative on a volume of storage or distribution medium does not bring the other program under the scope of these terms.
Tree2C
is a software developed in 2017-2021
by Alessandro Pedretti
All rights reserved.
Alessandro Pedretti
Dipartimento di Scienze Farmaceutiche
UniversitÓ degli Studi di Milano
Via Luigi Mangiagalli, 25
I-20133 Milano - Italy
Tel. +39 02 503 19332
Fax. +39 02 503 19359
E-Mail: info@vegazz.net
WWW: http://www.vegazz.net