12.12 The WarpEngine technology

|
12.12 The WarpEngine technology |
 |
Main topics:
The collaborative computing term includes technologies and informatics resources based on a network communication system that allows the documents and projects to be shared between users. All activities can be managed by a variety of devices such as desktops, laptops, tablets and smartphones. In a computational chemistry laboratory, one of the most common need of a researcher is not only to share information and data between the collaborators, but also computational resources for ab-initio calculations, semi-empirical calculations, MD simulations, virtual screening, data mining, etc.
The typical scenario of a computational chemistry laboratory is shown in the following scheme:
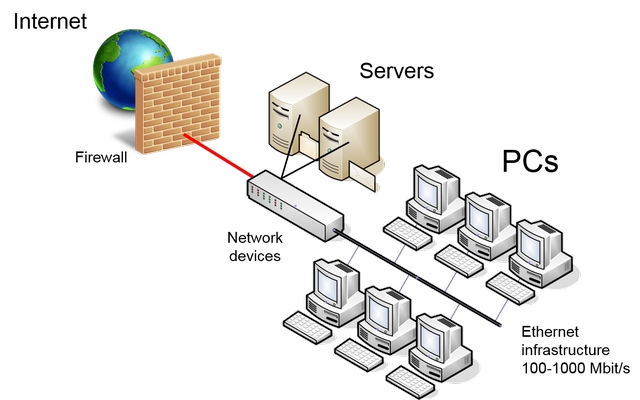
Several workstations are connected each other by network devices granting the
access to local resources, provided by one or more servers, and to Internet
trough a firewall. Therefore, the global computation power given by all PCs
taken together is very high, but is "fragmented" on the net, making hard the use
to run a single complex calculation. Moreover, the situation is more complex
by because the heterogeneity
of hardware and operating systems.
To overcome these problems, WarpEngine was developed whose main features are summarized here:
HTTP server Pages / sec. msec. / request WarpEngine 3 205.13 1.560 Internet Information Service 6.0 1 066.67 4.688 The benchmark was executed performing 100 requests with concurrency level of 5 using a four core CPU at 2.4 GHz.
These performances ensures on the same test system to extract (by SQL query), decompress and delivery molecules to the clients and receive the answer at the noticeable speed of 41 115.00 molecules / min with a peak performance of 79 651.78 jobs / min without the database management.
The security is provided by a tunnelling service and a digital signature that certify all files sent by the server to the clients.
As explained in the previous summary, WarpEngine is a client/server-based technology implemented in C++ language as part of the PowerNet plug-in and is fully integrated in the VEGA ZZ program. The same software can run as server or client, so you don't need specific versions for each personality. It can work indifferently in both LAN and WAN but, in this last case, some minor restrictions were introduced to grant the security.
The flowchart of the server side is depicted here:
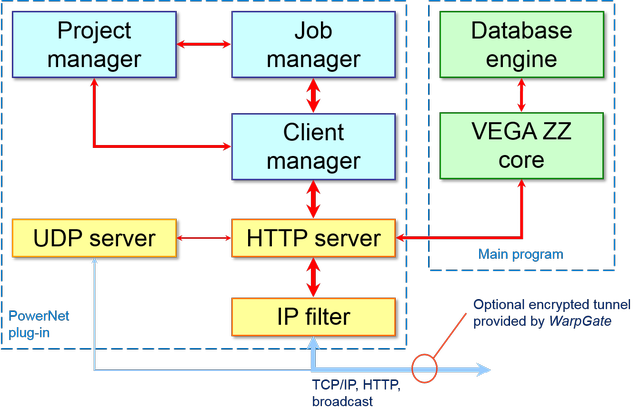
The server code includes several modules that are logically linked to each other. Due to the asynchronous nature of the code, some modules run as separated thread and the communication is implemented through events, mutexes and message queues that are provided by the cross-platform library named HyperDrive.
More in detail, here is the description of each module:
Project manager is one of the most important module at the server-side, as shown in the following scheme:
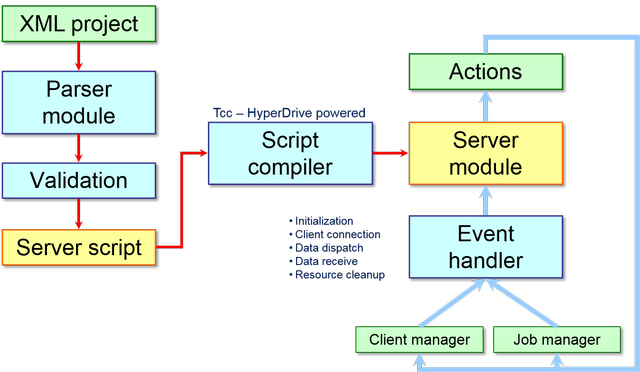
This server module plays a pivotal role because it does several important actions such as:
The client-side architecture is complementary if compared to that of the server-side as shown below:
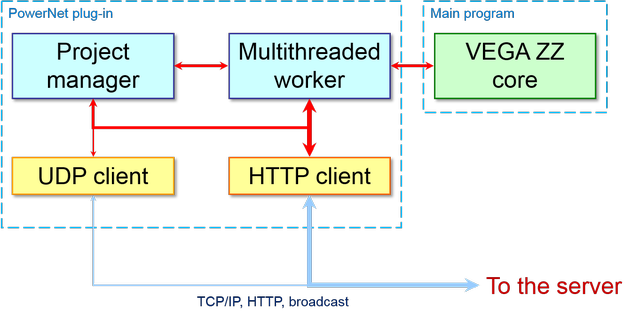
In particular, the most important client modules are:
Here is a more detailed representation of Project manager at the client-side:
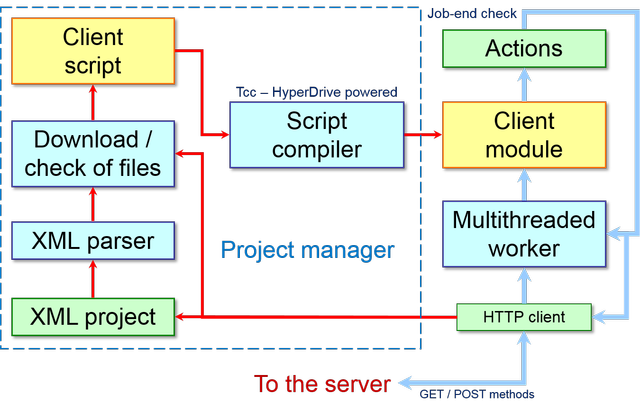
When the client connects itself to the server, several events occurs as shown below:
12.12.2 The graphic user interface
As explained above, VEGA ZZ includes both client and serve code, so when you start WarpEngine you can decide its running personality. If you want to test locally some applications, you can run two WarpEngine instances from two different VEGA ZZ on the same node, the former with the server personality and the latter with the client one.
12.12.2.1 The server personality
To start WarpEngine in server mode to setup a new calculation, you must
select File
![]() WarpEngine
WarpEngine
![]() Server: If your operating system is Windows and the
User Account Control (UAC) is enabled, administrative rights are required.
If they aren't available, VEGA ZZ asks you to restart the program to
obtain the rights.
Server: If your operating system is Windows and the
User Account Control (UAC) is enabled, administrative rights are required.
If they aren't available, VEGA ZZ asks you to restart the program to
obtain the rights.
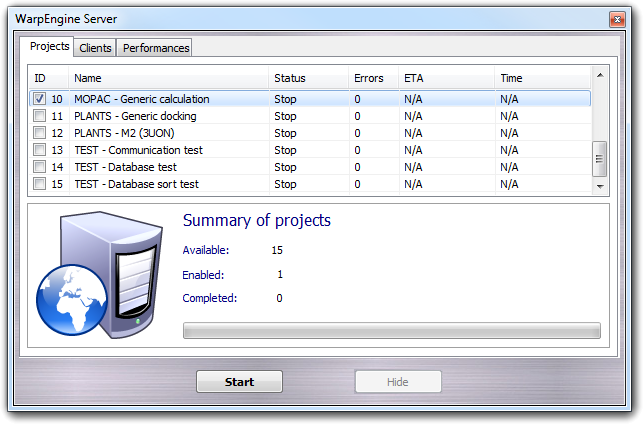
The main window show all available projects (see below to add a new one). In the Project tab, you can select the projects that you want to run. Although WarpEngine allows more than one project to be executed at the same time, there are calculations that needs the exclusive use of the resources and, for this reason, can be executed alone. For each project, the identification number (ID), the short description (Name), the status of the calculation (Status), the number of errors occurred during the calculation (Errors), the elapsed time to end the calculation (ETA) and the time spent by the calculation (Time) are shown as table. Status, Errors, ETA and Time are updated in real time when the project is running. In the Summary of projects box are shown the main information on the projects: the number of available projects (Available), the number of enabled/checked projects (Enabled) and the number of completed projects (Completed).
Clicking the Start button, the selected projects are executed. Additional data could be requested to the user according to the server script managing the calculation. For example, in same cases you must select the database to process or put some parameters, in other cases a full graphic interface is shown for the set-up of the calculation (e.g. PLANTS). When the set-up phase is completed, the server is ready for the incoming connection and you can minimize it on the system tray bar clicking Hide button.
Clicking the WarpEngine icon on the tray bar with the right mouse button, you can show its menu:
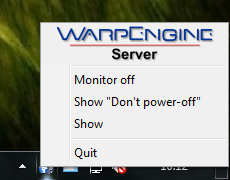
whose functions are summarized in the following table:
| Menu item | Description |
| Monitor off | Switch off the LCD monitor. |
| Show "Don't power off" | Show a large "WarpEngine - Don't power off" message on the desktop to prevent accidental power off by other users. You can hide the message double clicking it. |
| Show | Show VEGA ZZ and WarpEngine windows. |
| Quit | Stop WarpEngine server and close the program. A dialog box is shown to confirm the action. |
To stop the server without closing the program, you can click the Stop button: the action must be confirmed by a dialog box.
The Clients tab shows the connected clients, their status and some statistics about their activity:
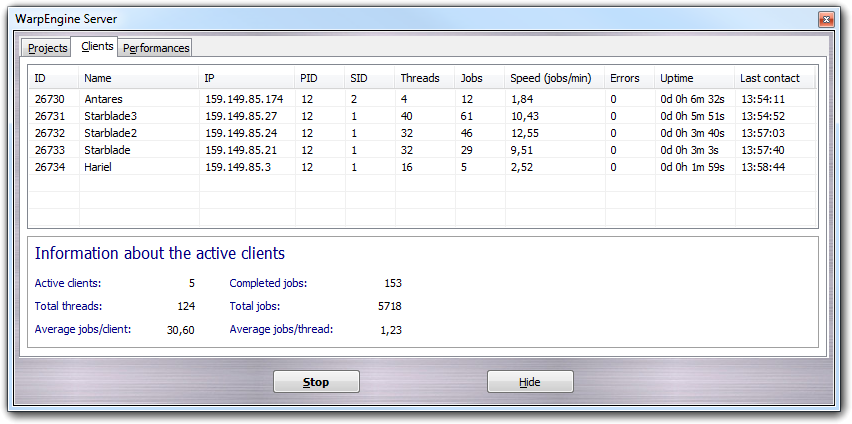
More in detail, for each client the identification code (ID), the host name (Name), the IP address (IP), the ID of the project on which the client is working (PID), the session ID (SID) used to identify different WarpEngine instances running on the same client, the number of working threads (Threads), the number of completed jobs (Jobs), the calculation speed expressed as number of jobs per minute (Speed (jobs/min)), the number or recoverable errors occurred, the time passed from the first contact (Uptime) and the hour of the last client contact (Last contact) are shown. Moreover, global information is reported at the bottom box: the number of active clients (Active clients), the number of completed jobs (Completed jobs), the total number of thread running at the same time (Total threads), the total number of jobs required to complete the calculation (Total jobs), the average number of jobs processed by each client (Average jobs/client) and the average number of jobs completed by each thread (Average jobs/thread).
After five consecutive errors, the client is automatically disconnected from the server and you can decide to disconnect manually the client by the context menu. By this menu, you can also recruit all client of the local network that are in waiting status. In this way, you don't need to connect manually each client to the server.
In Performances tab, you can monitor the performances of the calculation system in real time:
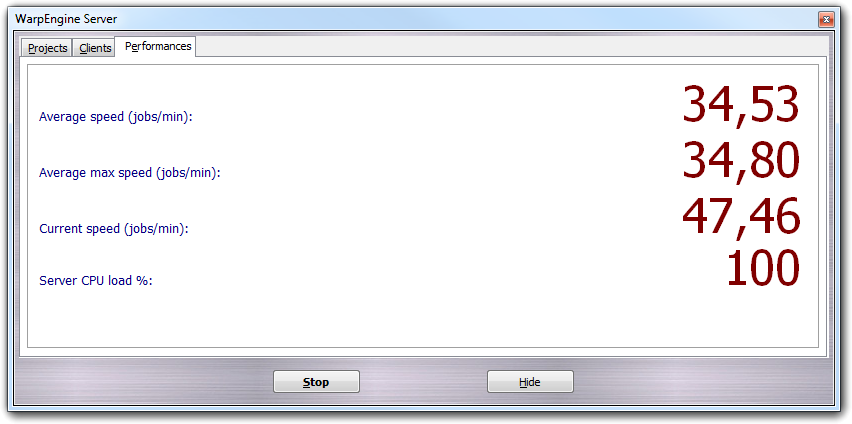
Here, you can check the average speed, the maximum average speed, the current speed (all these quantities are expressed as number of completed jobs per minute) and the CPU load of the server (as percentage).
12.12.2.2 The client personality
As for the server mode, to start WarpEngine with the client personality, you must
select File
![]() WarpEngine
WarpEngine
![]() Client. Also in this case, administrative rights are required. You can
decide to start the client and to connect it manually to a specific server or
leave it in a waiting status to be recruited by a server trough a broadcast
message. This second operative mode is available only if the server is in the
same network of the client or the client can access to the same local network
through a Virtual Private Network (VPN).
Client. Also in this case, administrative rights are required. You can
decide to start the client and to connect it manually to a specific server or
leave it in a waiting status to be recruited by a server trough a broadcast
message. This second operative mode is available only if the server is in the
same network of the client or the client can access to the same local network
through a Virtual Private Network (VPN).
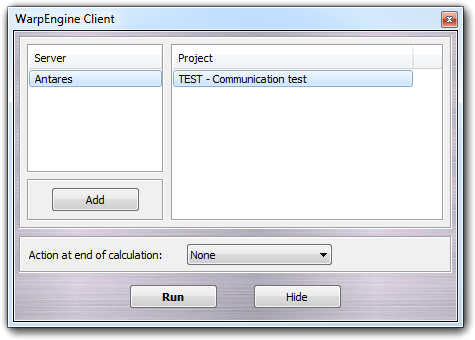
The servers are automatically detected if they are in the local network (Server column). Alternatively, they can be added manually clicking the Add button or choosing the Add item in the context menu:
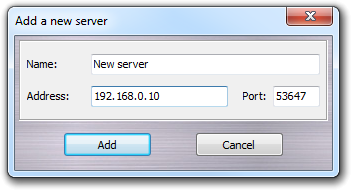
In this dialog window, you can specify the server name (Name, used only as alias), the IP address (Address) and the communication port (Port). Clicking Add, you can add the server to the list. The server is not saved and needs to be added every time that you need it.
Before to add the client to the calculation pool, you can choose the action performed at the end of the calculation: None (nothing is done), Close VEGA ZZ (closes the program) and Shutdown the system (power off the system).
To connect the client to the server and run the calculation, you must select the server and choose the project, thus click the Run button. The program is automatically minimized on the system tray showing a notification. You can minimize the client on the system tray without attaching it to a server just clicking the Hide button. In this way, the client remains in waiting mode and can be recruited by the server.
As for the WarpEngine server, if you click the icon on the system tray, you can show the control menu:
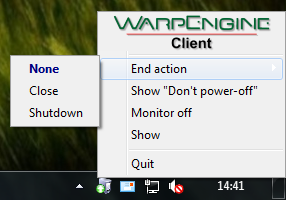
In addition, you can change the action performed at the end of the calculation, selecting the End action submenu as explained above.
12.12.3 WarpEngine pre-installed projects
VEGA ZZ package includes WarpEngine projects ready-to-use: some of them are for test purposes, while others are specific for intensive calculations.
These projects named with TEST prefix are suitable to test the network configuration, to check the client/server communication and to evaluate the transfer performances.
| Project name | Description |
| TEST - Communication test | This is a simple
stress-test to check the client/server communication in extreme
conditions. When you start the server, a file requester is shown to
choose the output file name that will be in CSV format, thus the project
asks you if you want to switch off all messages shown in VEGA ZZ
console. This function is useful to reduce the latencies an test the
full power of the network connection. The server send a packet of data to each client and collect the answers in the output CSV file. The test is repeated 10000 times and you can evaluate the speed in the Performances tab of the server. The output file includes four columns: the job number (JOBID, from 1 to 10000), the identification number of the client (CLIENTID), the identification number of the thread processing the message (THREADID, from 0 to n, where n is the maximum number of cores/threads minus 1 manageable by the client) and a progressive number as returning message (RESULT). |
| TEST - Database test | Also this project is a
stress test to check if the database functions implemented in WarpEngine
work properly. It requires a SQL database of molecules in any format
supported by VEGA ZZ (for more information,
click here). The server asks you
also for the output file name that will be written in CSV format. The server send a molecule to the client and receive a confirm message. If the operation is successfully completed the output file is updated. It includes two columns: the identification number of the molecule (JOBID) and the molecule name (MOLECULE). |
| TEST - Database sort test | This project is quite similar if compared to the previous one, but the main difference consists of how the output is written. The client downloads a molecule from the server, calculates its mass and returns this value as answer. The server sorts the results by mass and periodically (every 60 seconds) writes the output file. This file includes three columns: the job identification number (JOBID), the molecule name (MOLECULE) and the mass (MASS). |
12.12.3.2 APBS - Solvation energy
This project calculates the solvation energy (in water solvent) of a set of molecules in a database. It uses APBS (included in VEGA ZZ package) that is a software, developed by Nathan Baker in collaboration with J. Andrew McCammon and Michael Holst, for modelling biomolecular solvation through solution of the Poisson-Boltzmann equation (PBE).
When you start the project, you can select the input database and the output CSV file in which will be stored the following data:
| Column | Description |
| JOBID | Identification number of the job. |
| MOLECULE | Molecule name. |
| APBS_REF | Reference electrostatic energy in vacuum. |
| APBS_SOLV | Electrostatic energy in solvated state. |
| APBS_DIFF | Solvation energy calculated as difference between APBS_SOLV and APBS_REF. |
All energies are expressed in kJ/mol.
A log file is also created and named as the CSV one changing the extension from csv to log.
You can change the calculation parameters editing apbs.in file in ...\VEGA ZZ\Data\WarpEngine\Projects\APBS directory.
For more information on the implemented method, you can look here.
12.12.3.3 MOPAC - Semiempirical calculation
By this project, you can perform MOPAC calculations on molecules included into a database. MOPAC 7.01-4, that is included in VEGA ZZ package, is used by default but if a newer release is installed (click here for the installation guide), it is automatically detected and used. Don't mix different MOPAC versions: all clients must have the same MOPAC release.
When you start the server, you can select the input database including the molecules to process, the output CSV file in which the parameters calculated for each molecule are stored and the keywords required by MOPAC. The pre-defined keywords are AM1 PRECISE GEO-OK, while CHARGE keyword is not required, because the total charge is calculated automatically.
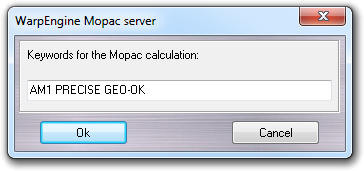
A new database is also created with the same format of the input database and with the same name of the CSV file adding - MOPAC suffix to store the structures optimized by MOPAC.
If any molecule is skipped during the calculation, a CSV file with the same name of the output file is created with skipped suffix in which the job identification code (JOBID) and the molecule name (MOLECULE) of the molecule with errors is saved.
If you stop the calculation accidentally or a heavy problem occurs to the server, you can restart the calculation just starting the server and setting up the calculation with the same parameters and files (including the output).
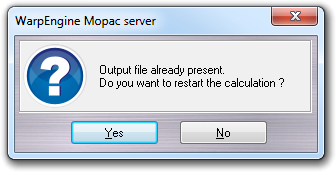
When you confirm to overwrite the output file a requester is shown to ask you if you want to restart the calculation.
The CSV output file includes the first two columns that are common to all calculation types: the identification number of the job (JOBID) and the molecule name (MOLECULE). Additional columns are added according to the specified MOPAC keywords and the relative values are retrieved from .arc files. If you add SUPER as keyword, the superdelocalizability data is read from the MOPAC output file.
12.12.3.4 PLANTS - Virtual screening
WarpEngine allows large scale virtual screening campaign to be performed in easy way by PLANTS software. This program must be installed at least on the server because the PLANTS server sends not only the data required for the calculation (protein target, control file and ligands to dock) but also the executable of the docking program. The executable is digitally signed in order to avoid the injection of malicious programs. To install PLANTS, you must follow the steps shown in the activation and installation section.
When you start WarpEngine with the server personality selecting PLANTS - Virtual Screening as project, a dialog to setup the calculation is shown:
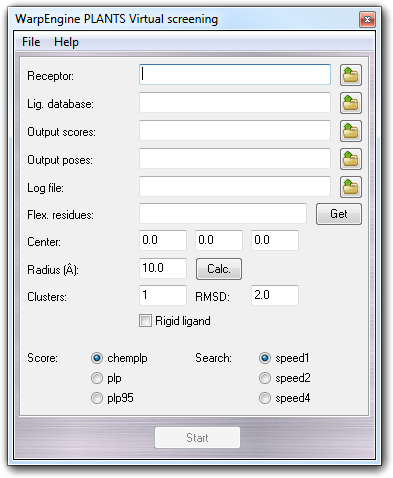
The setup procedure is quite similar to that for the normal PLANTS docking. More in detail, you can set:
File Description base_name.csv Best poses of each ligand ranked by lowest-to-highest total interaction energy (TOTAL_SCORE). base_name All.csv All poses of each ligand not ranked. base_name Mean.csv Mean, minimum, maximum, range and standard deviation values of each PLANTS score evaluated for all poses of a ligand (see Clusters). All these files are updated every 300 seconds and you can change this frequency editing WES_UPDFREQ constant in server.c file. If the number of clusters (Clusters) is set to 1, base_name All.csv and base_name Mean.csv have no additional meaning because include the same information of base_name.csv file.
Moreover, the menu bar allow other functions to be accessed as shown in the following table:
| Menu | Item | Shortcut | Description |
| File | Load config. | Ctrl+L | Load the settings from file. |
| Save config. | Ctrl+S | Save the settings to file. | |
| Quit | - | Close the set-up window and stop the project execution. | |
| Help | Manual | Ctrl+H | Show this manual. |
Clicking the Start button, the server is initialized and waits for incoming connections. As for MOPAC, If the output CSV file is already present by because a previous calculation was performed without finishing it, a dialog window asks you if you want restart the calculation or repeat it from the beginning.
This project is an example showing how to set up a PLANTS virtual screening without the use of the graphic user interface. More in detail, this project performs a docking calculation on M2 muscarinic receptpor (PDB ID 3UON). Pre-defined PLANTS projects are useful if you have to perform several docking studies on the same target keeping the same parameters.
To build your own pre-defined project you must follow these steps:
Restarting VEGA ZZ, your project will appear in the main window of WarpEngine server. When you run the project, two file requesters are shown to choose the ligand database and the base file name of the CSV outputs. The restart feature is also available as for calculations prepared by graphic user interface.
RESCORE+ is a WarpEngine project that allows the calculation of different scores from a given set of poses obtained by a previous docking simulation. It is also possible to perform the same calculation using a trajectory file as input in order to evaluate how the interaction between two molecules (usually a receptor and a generic ligand) evolves during the molecular dynamics simulation.
RESCORE+ accepts different input combinations:
The files must be in one of the file/database formats supported by VEGA ZZ (see supported file formats, supported database formats). If you provide files including both ligand and receptor (such as a single database or MD trajectory), the ligand must be le last molecule in the molecular assembly in order to be correctly detected.
Before to calculate the scores, the complexes can be minimized by NAMD 2 (that must be installed at least on the WarpEngine server), applying spherical constraints around the ligand if needed by the user. Moreover, the processed complexes can be saved as single files (you can choose the file format) or collected in a whole database (this is the best choice if you need to process a large number of molecules).
When you start WarpEngine, with the server personality selecting RESCORE+ - Rescoring of docked poses as project, a dialog to setup the calculation is shown:
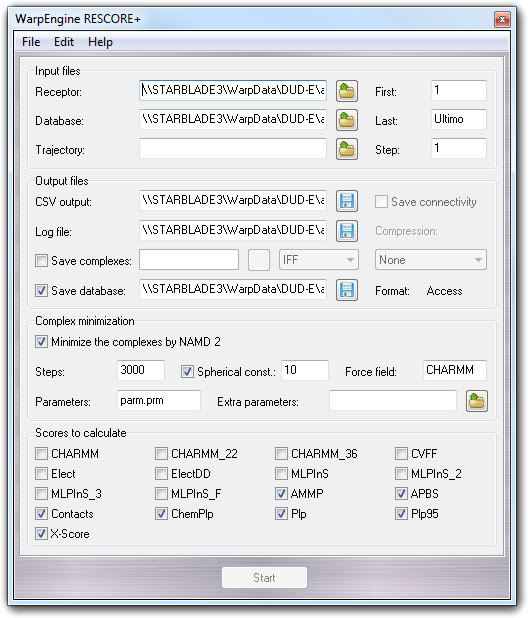
In the Input files box, you can choose the receptor file or the complex file if you want to rescore a trajectory (Receptor). This field could be empty if the database doesn't include the ligand poses but the whole ligand-receptor complexes.
You can also set the trajectory file (Trajectory) if you are rescoring the output of a MD simulation, the first (First) and the last (Last) pose/frame number and the pose/frame increment (Step) if you want to process only a part of the set of poses/frames.
In the Output files box, you can select the file in which are stored the calculated score for each pose/frame (CSV output). This file is in Comma Separated Values format (CSV) that is read by the most popular spreadsheet programs. Furthermore, you can select the log file (Log file) in which are recorded the calculation settings and the possible error messages. Checking Save complexes, you enable the capability to save the ligand-receptor complexes in a specific directory. More in detail, you can specify also the file format, the compression algorithm (Compression) and if to save the connectivity for PDB files (Save connectivity).
Checking Save database, the complexes are collected in a database that you can specify by the file requester.
In the Complex minimization box, you can find the parameters required for the complex minimization. Checking Minimize the complexes by NAMD 2, you can enable this feature and in particular you can set the number of iterations (Steps), the force field (Force filed), the parameter file (Parameters) that must be in ...\VEGA ZZ\Data\Parameters directory and the optional extra parameter file (Extra parameters) that is used to add parameters not included in the main file. Checking Spherical const., only the atoms included included in a sphere of a given radius (e.g. 10 ┼) centred on the ligand barycentre are free to move and the others out of this sphere are kept fixed.
In the Scores to calculate box, you can choose the scores to consider in the rescore procedure as shown in the following table:
| Score | Type | Program |
Included in VEGA ZZ |
Description |
| AMMP | External | AMMP | Yes | Electrostatic and Van der Waals interaction calculated by SP4 force field. |
| APBS | External | APBS | Yes | Electrostatic interaction evaluated by Adaptive Poisson-Boltzmann Solver (APBS). |
| CHARMM | Internal | - | Yes | R6-R12 non-bond interaction evaluated by CHARMM 22 force field provided by Accelrys. To perform this calculation, the parm.prm file must be copied in the ...\VEGA ZZ\Data\Parameters directory. This file is not included in the package for copyright reasons. |
| CHARMM_22 | Internal | - | Yes | R6-R12 non-bond interaction evaluated by CHARMM 22 force field. |
| CHARMM_36 | Internal | - | Yes | R6-R12 non-bond interaction evaluated by CHARMM 36 force field. |
| CVFF | Internal | - | Yes | R6-R12 non-bond interaction evaluated by CVFF force field. |
| ChemPlp | External | PLANTS | No | Knowledge score implemented in PLANTS software. |
| Contacts | WarpEngine | - | Yes | Score that evaluates the number of contacts between the interaction partners. |
| Elect | Internal | - | Yes | Electrostatic interaction based on Coulomb equation. The dielectric constant is set to 1.0. |
| ElectDD | Internal | - | Yes | Distance-dependent electrostatic interaction based on Coulomb equation. The dielectric constant is set to 1.0. |
| MLPInS | Internal | - | Yes | Hydrophobic interaction calculated using the Broto's and Moreau's atomic constants. |
| MLPInS2 | Internal | - | Yes | Hydrophobic interaction in which the distance between interacting atom pairs is considered as square value. |
| MLPInS3 | Internal | - | Yes | Hydrophobic interaction in which the distance between interacting atom pairs is considered as cube value. |
| MLPInSF | Internal | - | Yes | Hydrophobic interaction in which the distance is evaluated by the Fermi's equation. |
| Plp | External | PLANTS | No | Knowledge score implemented in PLANTS software. |
| Plp95 | External | PLANTS | No | Knowledge score implemented in PLANTS software. |
| X-Score | External | X-Score | No | Binding score evaluated by X-Score program. |
For more information about MLPInS, click here.
Clicking the Start button, the WarpEngine server is initialized and becomes ready for incoming connections.
Other functions are available through the menu bar as shown in the following table:
| Menu | Item | Subitem | Shortcut | Description |
| File | Load config. | - | Ctrl+L | Load the settings from a configuration file. |
| Save config. | - | Ctrl+S | Save the current settings to a configuration file. | |
| Quit | - | - | Close the configuration dialog window. | |
| Edit | Select scores | Default | - | Select the default scoring functions. |
| All | - | Select all scoring functions. | ||
| None | - | Unselect all scoring functions. | ||
| Clear config. | - | - | Clear all configuration fields and revert to default. | |
| Help | Manual | - | Ctrl+H | Show this manual. |
Notes:
to enable all functions of this WarpEngine project, you must install the following external programs/files:
You don't need to install these optional components on the clients, because WarpEngine sends them when a new worker is added to the calculation pool.
12.12.4 Developing a new WarpEngine application
The general purpose WarpEngine core can be adapted to specific calculations through server and client scripts. As explained above, they must be written in C-Script language that allows the direct access to WarpEngine and HyperDrive APIs.
The WarpEngine files are located in ...\VEGA ZZ\Data\WarpEngine
directory whose full path changes on the basis of operating system version. To find it in
easy way, you can type OpenDataDir in VEGA Console (command prompt) or
select Help
![]() Explore data directory in VEGA ZZ menu bar.
Explore data directory in VEGA ZZ menu bar.
Here is the typical directory tree:
| VEGA ZZ | ||||
| Config | ||||
| Data | ||||
| WarpEngine | ||||
| Projects | ||||
| Project_1 | ||||
| Project_2 | ||||
| Project_n | ||||
| Templates | ||||
| Template_1 | ||||
| Template_2 | ||||
| Template_n | ||||
| Scripts | ||||
The Projects directory includes the data for each project that must be placed in sub-directory (e.g. Project_1), while Templates directory contains the auxiliary files that can be used without changes in different projects.
In each project directory, project.xml file must be present to describe the calculation and the following scheme shows the meaning of each XML tag:
|
Configuration file |
Description |
|
<weproject version="1.0" enabled="0" multithread="1" runalone="1"> |
Main tag:
|
|
<startuplogo file="%WEPRJECTSDIR%/Project_1/logo.png" delay="2000" fadespeed="250"> |
Splash logo shown at start-up (optional):
|
|
|
|
||
<name lang="english">Project title</name> |
Project name. This tag supports the language localization by lang option and therefore it can be present one time fore each language that you want to support:
|
|
<description lang="english">
Description of the project
</description>
|
Description of the project. Also in this case, the description can be localized by lang option:
|
|
<arch>ALL</arch> |
Type of hardware architectures supported by the project: ALL (= all supported by WarpEngine), LINUX_ARM, LINUX_X64, LINUX_X86, WIN_ARM, WIN_X64 and WIN_X86. |
|
|
|
||
<client src="%WETEMPLATEDIR%/Template_1/client.c"/> |
Client script (optional). It must be specified if the client script is
not in the project directory:
|
|
|
|
||
<server src="%WETEMPLATEDIR%/Template_1/server.c"/> |
Same of above but for the server. This tag is optional: if it's missing, it will be used the server.c file placed in the project directory. |
|
<cliramdisk enabled="1" basesize="20" threadsize="1"/> |
Client ram disk used for the temporary files. It is a block of ram used as disk drive to improve the I/O performances and to preserve the integrity of drives that are sensible to continuous read/write operations such as SSDs. The ram disk is provided by ImDisk driver that must be installed on the client.
WARNING: If ImDisk is not installed, the calculation runs anyway and the temporary files are created in the system disk. A warning message is shown. |
|
<srvramdisk enabled="1" size="20"/> |
It is the same of above, but for the server side.
|
|
|
|
||
<download> |
Begin of the download section in which the files needed by the client must be specified. | |
<file src="client.c"
map="%WETEMPLATEDIR%/Template_1/client.c"
unzip="0"/>
<file src="data.zip"
map="%WETEMPLATEDIR%/Template_1/data.zip"
dest="%WorkDir%" unzip="1"/>
|
File to download:
|
|
</download> |
End of download section. |
|
</weproject> |
End of the project configuration file. |
|
The empty project (see ...\Projects\EMPTY directory) is a good starting point to build a new calculation project. It includes a pre-defined project.xml file in which you can define the main parameters as explained above and client.c and server.c scripts. Both scripts supports the language localization, includes different functions called when a specific event occurs and use extensively the HyperDrive library.
To modify these scripts, you must have basic skills in C programming (C-Scripts are fully C-99 compliant).
Both client and server scripts can access to global variables declared in .../VEGA ZZ/Tcc/include/vega/vgplugin.h, .../VEGA ZZ/Tcc/include/vega/wengine.h and .../VEGA ZZ/Tcc/include/vega/wetypes.h
This structure includes
information on VEGA ZZ and allows the access to their super APIs. The
complete description of the structure is available in the
plug-in SDK section.
This structure is useful to retrieve the information on the project and it so defined:
typedef struct {
HD_LONG Size; /* Structure size for versioning */
HD_LONG * PrjStatus; /* Project status pointer */
HD_LONG * SrvPort; /* Server TCP/IP port */
HD_STRING PrjDesc; /* Project description */
HD_STRING PrjName; /* Project name */
HD_STRING PrjPath; /* Project path */
HD_STRING PrjURL; /* Project URL */
HD_STRING Server; /* Server name or IP */
HD_STRING TmpPath; /* Path of the temporary directory */
HD_ULONG * PrjID; /* Project ID */
HD_ULONG Threads; /* Number of threads */
} WE_WEINFO;
12.12.4.2 Events managed through the server script
This script implements the actions given as answer for a specific event at server-side. In the following flowchart are summarized the event handled by the server script:
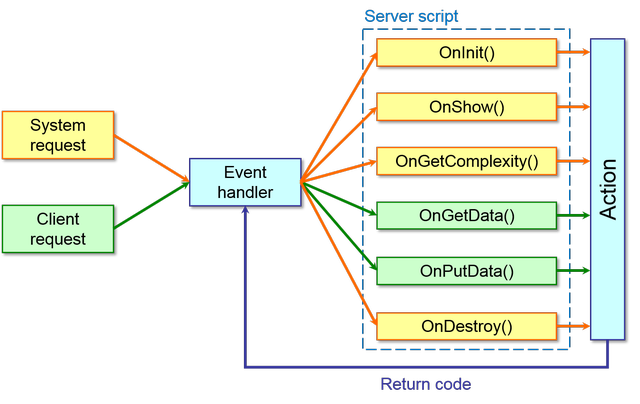
The system and the client requests are dispatched by the event handler calling the specific functions of the server script that implement user-defined actions. Each event function return a code to inform the Event handler if the action successes or not.
More in detail, the event are managed by the following functions:
This function is called during the initialization phase of the server to obtain the complexity (size) of the calculation. In other words, this function must return the total number of jobs (JobTot) and a vector of bytes with JobTot size used by the server to track each job. Remember to release the vector in OnDestroy() function.
In server.c of EMPTY project, the code to get the complexity of a
database of molecules is already implemented as example.
This event occurs when the client ask the server for the data required to complete a single job. For example, it can be the URL to retrieve the molecule that mast be processed. The returned data is a string list (StrList) that can be populated by HD_StrListAdd() HyperDrive function (for more information see hdstring.h header in ...\VEGA ZZ\Tcc\include\hyperdrive directory). JobID and SrvIP are respectively the identification number of the job and the IP address of the server.
This function must return TRUE if no error occurs, otherwise FALSE.
This is the initialization code called one time only when the server is started. For example, here you can open the database containing the molecules that must be processed during the calculation. Also for this function, all allocated resources can be freed in OnDestroy().
The return value must be TRUE or FALSE if everything is ok or an error
occurs during the initialization.
This function is called
every time that the client send the results encoded in a string list (StrList)
and/or in raw binary format (Data pointer of DataSize size).
JobID is the identification code of the job sending the data and
JobStatus is useful to retrieve if the job was completed or some errors
occurs. The complete list of job status codes is in ...\VEGA ZZ\Tcc\include\vega\wetypes.h
header.
This event handler is optional and is called to show the custom graphic user interface that can be used to configure the calculation. PLANTS project is an example of a server script that uses GraphAppZ APIs for the graphic interface. Flags argument is not used and is present for future implementations.
To abort the calculation at set-up phase, this function must return FALSE, otherwise TRUE informs the server to continue to the next initialization phase.
12.12.4.3 Events managed through the client script
Also the client script can handle several events that are depicted in the following scheme:
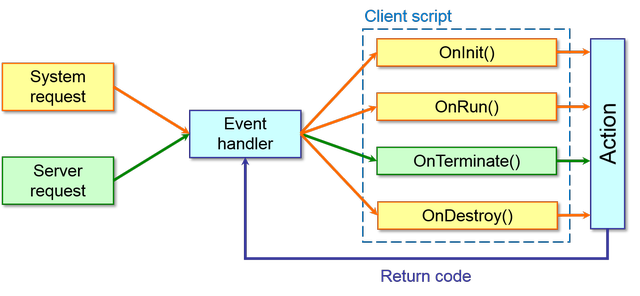
In particular, here is the description of each event function:
This function is called after the initialization phase when the connection with the server is established. The arguments are the identification codes of the thread (ThreadID) and the client (ClientID). This section of code must include the calculation loop which at every step, download the data from the server, process it and upload the results to the server. Several specific API, explained below, helps you to implement these actions.
The function must return TRUE or FALSE if the calculation is successfully
completed or an error occurs.
WARNING:
This function is called by multiple threads according to the number of
thread manageable by the hardware. It means that if you need the exclusive
access to specific resources by a single thread, you must use the functions
of a thread messaging system such as barriers, mutexes, semaphores, etc.
HyperDrive library includes OS-independent APIs for thread messaging
(see ...\VEGA ZZ\Tcc\include\hyperdrive\hyperdrive.h header).
This function is called when the calculation is killed externally by the server or directly by the user. You can put here the code to signal that the calculation threads must abort their activity as soon as possible. The best way to do it, it to call WecRunSignal(FALSE) function (see below).
The prototypes of WarpEngine APIs are defined in .../VEGA ZZ/Tcc/include/vega/wengine.h header.
APIs for both client and server scripts:
HD_LONG VegaCmdEx(const HD_CHAR *Com, ...)
HD_LONG VegaCmdSafe(HD_STRING ResStr, const HD_CHAR *Com, ...)
Send a command (Com) to VEGA ZZ. For a complete list of the command, you can refer to the script section. If the command fails, a non-zero value returned according to the error code. VegaCmdEx allows the format expansion as the printf() command, while VegaCmdSafe is the thread-safe version the returns the command result in the ResStr HyperDrive string (that must be previously initialized). ResStr can be NULL if you don't need to retrieve the command result. The non thread-safe versions put the command result in VgInfo -> Result (for more details, see below).
Due to the multithreading architecture of WarpEngine, it is strongly
recommended to use VegaCmdSafe() instead of the other versions.
Load a molecule from the memory in VEGA ZZ environment. This function is very useful to avoid the I/O overload. In a typical scenario, the client get the molecule to process from the server and receive it a memory segment. It the molecule must be modified or converted to another format, you must load it in VEGA ZZ, but the Open and OpenEx commands can load data only from files and URLs.
The input are the pointer of the memory segment including the molecule in any format supported by VEGA ZZ (MemPtr), the size of the memory segment (Size) in bytes and the load flags (Flags) that can be VG_LOADMEM_NONE or VG_LOADMEM_SILENT (no messages are shown in VEGA ZZ console during the loading).
If the molecule is correctly loaded in VEGA ZZ, the function returns TRUE, otherwise if an error occurs, it returns FALSE.
Print a message in VEGA ZZ console. It works exactly as printf(), but the output is redirected to VEGA ZZ console. Moreover, it implements %a formatting to print HD_STRING without any conversion. For a complete list of the format specifiers, click here.
Decode a XML reply. All client-server messages are XML encoded, so you need to parse them. WarpEngine includes MXML library that is automatically linked to the scripts to help you in this operation. WeDecodeXmlReply() is a high level implementation of MXML library that allow to obtain the XML tree in easy way. The input is a string including the XML text and optionally the Status and ErrCode pointers accepting main message data. The message can be decoded by mxmlFindElement() function:
#include <wengine.h>
HD_ULONG JobID; mxml_node_t * Node; mxml_node_t * Tree;
...
/*** Data includes the XML reply ***/
if ((Tree = WeDecodeXmlReply(Data, NULL, NULL)) != NULL) {
Node = Tree;
while((Node = mxmlFindElement(Node, Tree, "data", "id", NULL, MXML_DESCEND)) != NULL) {
switch(atoi(mxmlElementGetAttr(Node, "id"))) {
case 1: /* Job ID */
sscanf(Node -> child -> value.opaque, "%u", &JobID);
break;
case 2: /* URL of the ligand to download */
HD_StrCpyC(Data, Node -> child -> value.opaque);
break;
} /* End of switch */
} /* End of while */
mxmlDelete(Tree);
}
Run an external program. The parameters are:
CmdLine
![]() Command line to run the program.
Command line to run the program.
FilePath
![]() Path of the program executable.
Path of the program executable.
StartMsg
![]() Message shown in VEGA ZZ console (optional).
Message shown in VEGA ZZ console (optional).
Info
![]() Process information pointer (optional).
Process information pointer (optional).
Flags
![]() Start-up flags (reserved for future implementations). Set it to 0.
Start-up flags (reserved for future implementations). Set it to 0.
APIs only for client scripts:
Change the status of the
client from stopped to running (TRUE) and vice versa (FALSE).
This function is useful to send some data to the server such as the results of a calculation. You must specify the client identification code (ClientID), the job identification code (JobID) the job status (JobStatus) and the string containing the data (Data). ClientID is passed through OnRun() event, JobID is obtained decoding the server answer when you ask for the next job and JobStatus can assume different values according .../VEGA ZZ/Tcc/include/vega/wetypes.h header.
The return value can be TRUE or FALSE if the data transmission occurs without errors or not.
Notify a non-recoverable error to the server. If the notification fails, FALSE is returned.
Several macros are defined in .../VEGA ZZ/Tcc/include/vega/wengine.h header in oder to help you in script programming.
HD_DLONG a = 10;
printf("64 bit integer: %" _FMT64 "d\n", a);
HD_ATOM *Atm;
for(Atm = VegaFisrtAtom; Atm; Atm = Atm -> Ptr) {
/* Put here your code */
}
12.12.4.6 Minimalist server code
This section shows the minimalist server script that you can use to process a database of molecules. It is a simplified version of EMPTY project
/*********************************
**** WarpEngine Server Script ****
*********************************/
/* Save this file as ...\VEGA ZZ\Data\WarpEngine\Projects\Example\server.c */
/* WarpEngine header */
#include <wengine.h>
/* HyperDrive headers */
#include <hdargs.h>
#include <hddir.h>
#include <hdtime.h>
/* Standard C headers */
#include <string.h>
#include <stdio.h>
#include <stdlib.h>
/**** Global variables ****/
HD_BOOL Running = TRUE; // Flag to control the server activity (FALSE = stop it, boolean variable)
HD_BYTE * StatVect = NULL; // Vector to monitor the job activities (see OnGetComplexity(), byte pointer)
HD_STRING hDbIn = NULL; // Handle of the database to process (HyperDrive string)
HD_STRING DbInName = NULL; // Name of the database to process (HyperDrive string)
HD_UDLONG NumJob = 0LL; // Number of jobs to complete (64 bit unsigned integer)
/**** Show an error ****/
HD_VOID Error(const HD_CHAR *Err)
{
/* Uses VEGA ZZ MessageBox to show the error */
VegaCmdSafe(NULL, "MessageBox \"%s.\" \"ERROR\" 16", Err);
}
/**** Destroy event ****/
HD_BOOL OnDestroy(HD_VOID)
{
Running = FALSE; // Set the run flag to FALSE
/*
* Put here your code
*/
/* Free the resources */
HD_FreeSafe(StatVect); // Free the vector to monitor the jobs
HD_StrFree(DbInName); // Free the HyperDrive string of database name
/* Close the database */
if (!HD_StrIsEmpty(hDbIn)) { // Check if the handle is not empty
VegaCmdSafe(NULL, "DbLock %a 0", hDbIn); // Unlock the database
VegaCmdSafe(NULL, "DbClose %a", hDbIn); // Close the database
}
/* Print into VEGA ZZ console */
VegaPrint("* Server terminated\n");
return TRUE;
}
/**** Get the complexity ****/
HD_BYTE * OnGetComplexity(HD_UDLONG *JobTot)
{
HD_STRING Res = HD_StrNew(); // Create a new HyperDrive string variable
/* Ask to VEGA how many molecules are in the database */
if (!VegaCmdSafe(Res, "DbInfo %a Molecules", hDbIn)) { // If it falis,
Error("Unable to get the number of molecules"); // the error message is shown,
HD_StrFree(Res); // the Res string is freed
return NULL; // and the function return a NULL pointer (error condition)
}
/* Convert the string to a 64 bit unsigned integer */
NumJob = HD_Str2ULint(Res);
HD_StrFree(Res); // Res is no more needed, so it is freed
if (NumJob == 0LL) { // If NumJob is 0 (the database is empty),
Error("Empty database"); // the error message is shown
return NULL; // and the function return a NULL pointer (error condition)
}
VegaPrint("* Molecules in database: %llu\n", NumJob); // Print how many molecules are in the database
/**** Allocate the byte vector to track the jobs ****/
StatVect = (HD_BYTE *)HD_Calloc(NumJob); // HD_Calloc() initilizes the vectot to 0
if (StatVect == NULL) { // If the vector pointer is NULL,
Error("Out of memory"); // the error message is shown
return NULL; // and the function return a NULL pointer (error condition)
}
*JobTot = NumJob; // Put the number of jobs
/*
* Put here the user code
*/
/* Show waiting message */
VegaPrint("* Waiting for incoming connections ...\n\n");
/* Return the job vector poiner */
return StatVect;
}
/**** Get the data for the job ****/
HD_BOOL OnGetData(HD_STRLIST StrList, HD_UDLONG JobID, HD_ULONG SrvIP)
{
HD_STRING IpStr = HD_StrNew(); // Create new HyperDrive string variables
HD_STRING DbURL = HD_StrNew();
/* Convert the IP V4 address to string */
HD_StrFromIPV4(IpStr, SrvIP);
/* Create the URL to get the molecule */
HD_StrPrintC(DbURL, "http://%a:%d/dbase.ebd?Cmd=getmol&Db=%a&RowID=%llu",
IpStr, // Server IP address
*WeInfo -> SrvPort, // Server IP port
DbInName, // Database name
JobID); // Job ID = Molecule ID to download
/* Put the URL string in the the answer list */
HD_StrListAdd(StrList, DbURL);
/* Free the resources */
HD_StrFree(IpStr);
HD_StrFree(DbURL);
/* Show the message */
VegaPrint("* Sending job %llu\n", JobID);
return TRUE;
}
/**** Initialization code ****/
HD_BOOL OnInit(HD_VOID)
{
/* Starting message */
VegaPrint("\n**** WarpEngine Server Example ****\n\n");
/* Ask for the input database */
DbInName = HD_StrNew();
VegaCmdSafe(DbInName, "OpenDlg \"Open input database ...\" \"\" \"All supported databases|*.accdb;*.db;*.dsn;*.mdb;*.sdf\" 1");
if (HD_StrIsEmpty(DbInName)) // If the database name is empty (cancel or no selection),
return FALSE; // the function return FALSE, forcing the exit
/* Open the database */
hDbIn = HD_StrNew(); // Create a new string variable in which to put the database handle
VegaCmdSafe(hDbIn, "DbOpen \"%a\"", DbInName); // Open the database and put in hDbIn the handle
if (*hDbIn -> Cstr == '0') { // If the database handle is 0,
Error("Can't open the database"); // the error message is shown
return FALSE; // and the function return FALSE, forcing the exit
}
/* Remove the path from the database file name */
HD_StrFileName(DbInName);
/* Lock the database to prevent the accidental close */
VegaCmdSafe(NULL, "DbLock %a 1", hDbIn);
/*
* Put here other initialization code
*/
VegaPrint("* Server initialized\n");
return TRUE;
}
/**** Receive the data from the client ****/
HD_BOOL OnPutData(HD_STRLIST StrList, HD_UDLONG JobID, HD_LONG JobStatus,
HD_VOID *Data, HD_ULONG DataSize)
{
HD_STRING Line = HD_StrNew(); // Create new HyperDrive string variables
HD_STRING Molecule = HD_StrNew();
/* Get the molecule name from database */
VegaCmdSafe(Molecule, "DbMolName %a %llu", hDbIn, JobID - 1LL);
if (HD_StrIsEmpty(Molecule)) // If the molecule name is unknown,
HD_StrCpyC(Molecule, "Unknown"); // set Molecule to "Unknown"
else // otherwise
HD_StrTrimRight(Molecule); // trim the string removing spacing and tabs
/*
* Put here the code to manage the results
*/
VegaPrint("* Results from job %llu (%a) received\n", JobID, Molecule);
/* Free the resources */
HD_StrFree(Line);
HD_StrFree(Molecule);
return Running;
}
12.12.4.7 Minimalist client code
This section shows the minimalist client script that is a simplified version of EMPTY project
/*********************************
**** WarpEngine Client Script ****
*********************************/
/* Save this file as ...\VEGA ZZ\Data\WarpEngine\Projects\Example\client.c */
/* WarpEngine header */
#include <wengine.h>
/* HyperDrive headers */
#include <hdsocket.h>
#include <hdtime.h>
/* MXML header */
#include <mxml.h>
/* Standard C headers */
#include <stdio.h>
#include <stdlib.h>
#include <time.h>
/**** Global variables ****/
HD_MUTEX MtxVega = NULL; // HyperDrive mutex handle for exclusive access to VEGA ZZ resources (handle)
HD_UDLONG JobCount = 0LL; // Job counter variable (64 bit unsigned integer)
HD_ULONG Threads = 0; // Number of threads (32 bit unsigned integer)
/**** Show an error ****/
HD_VOID Error(const HD_CHAR *Err)
{
/* Uses VEGA ZZ MessageBox to show the error */
VegaCmdSafe(NULL, "MessageBox \"%s.\" \"ERROR\" 16", Err);
}
/**** Destroy event ****/
HD_BOOL OnDestroy(HD_VOID)
{
HD_MthCloseMutexEx(MtxVega); // Close the mutex
/*
* Put here your code
*/
return TRUE;
}
/**** Initialization code ****/
HD_BOOL OnInit(HD_VOID)
{
/* Starting message */
VegaPrint("\n**** WarpEngine Client Example ****\n\n");
/* Create the mutex */
MtxVega = HD_MthCreateMutexEx();
/*
* Put here other initialization code
*/
/* Set the client status to running */
WecRunSignal(TRUE);
/* Show the message that the initialization is OK */
VegaPrint("* Client initialized\n");
return TRUE;
}
/**** Run the job ****/
/*
* This function is called by each thread whose number depends
* on the number of cores and is the hyper-threading is available.
* For example, if the client node has 2 CPUs with 8 cores each and
* hyper-threading enabled (we assume 2 threads for each core),
* there are:
* 2 * 8 * 2 = 16 threads
* that run concurrently OnRun()
*/
HD_BOOL OnRun(HD_ULONG ThreadID, HD_ULONG ClientID)
{
HD_HTTPCLIENT DataClient; // Handle of HyperDrive HTTP client
HD_ULONG JobID; // ID of the running job
mxml_node_t * Node; // Node pointer of XML structure
mxml_node_t * Tree; // Tree pointer of XML structure
HD_STRING Data = HD_StrNew();
/* Trace the number of threads */
HD_MthMutexOnEx(MtxVega); // The mutex activation allows the exclusive access to code (only one thread at time)
++Threads; // Increment the total number of threads
HD_MthMutexOffEx(MtxVega); // Disable the exclusive access of the code
/* Create a new HTTP client */
DataClient = HD_HttpClientNew();
if (DataClient == NULL) // If it is impossible to do it,
goto ExitErr; // jump to ExitErr release the resources
/* Set the URL (Data) to get the data for the job */
HD_StrPrintC(Data, "http://%a:%d/getdata.ebd?ID=%u&ProjectID=%u",
WeInfo -> Server, // IP address of the server
*WeInfo -> SrvPort, // Port of the server
ClientID, // Client ID
*WeInfo -> PrjID); // Project ID
if (!HD_HttpClientParseUrl(DataClient, Data)) // Parse the URL, but if it fails,
goto ExitErr; // jump to ExitErr to release the resources
/* Here is the main loop for calculation */
while(WecIsRunning()) { // Check if the status is "run"
/* Get the data for the job */
HD_HttpClientGetStr(DataClient, Data); // Download from the server the XML data of the job
if (HD_StrIsEmpty(Data)) // If Data is empty (an error is occurred),
break; // the main loop exits
/* Parse the XML data */
Tree = WeDecodeXmlReply(Data, NULL, NULL); // Obtain the pointer of the XML tree
if (Tree != NULL) { // If the XML data is consistent ...
/* Decode the data for the job */
JobID = 0; // Initialize the variables
Node = Tree;
while ((Node = mxmlFindElement(Node, Tree, "data", "id", NULL, MXML_DESCEND)) != NULL) {
if (atoi(mxmlElementGetAttr(Node, "id")) == 1) {
sscanf(Node -> child -> value.opaque, "%u", &JobID);
break;
}
} /* End of while */
mxmlDelete(Tree); // Free the XML tree
/* Check if the calculation is finish */
if (!JobID) break;
/* Show the progress */
VegaPrint("* Running job %u\n", JobID);
/*
* Put here your code to perform the calculation
*/
/* Send the results */
HD_MthMutexOnEx(MtxVega);
HD_StrPrintC(Data, "&CLIENTID=%u&THREADID=%u&RESULT=%llu", ClientID, ThreadID, ++JobCount);
HD_MthMutexOffEx(MtxVega);
if (!WecSendData(ClientID, JobID, WE_JOBST_DONE, Data)) break;
}
} /* End of while */
/* Release the resources */
ExitErr:
HD_HttpClientFree(DataClient); // Free the HTTP client
HD_StrFree(Data); // Free the Data string
HD_MthMutexOnEx(MtxVega); // Enable the exclusive access to the code
--Threads; // Decrease the number of working threads
if (Threads == 0) // If there are no more working threads,
VegaPrint("* Calculation terminated\n"); // the calculation is terminated
HD_MthMutexOffEx(MtxVega); // Disable the exclusive access to the code
return TRUE;
}
/**** Terminate event ****/
HD_BOOL OnTerminate(HD_VOID)
{
WecRunSignal(FALSE); // Set the client status to stop
return TRUE;
}